2.2 Biochemistry and Cell Biology
KEY CONCEPTS
By the end of this section, you will be able to do the following:
- Explain the role of biochemistry in characterizing cell function, helping make the link between cell structure and function
- Evaluate the importance of the discovery of enzymes for cell biology
- Describe techniques that are useful for isolating cellular components, including molecules and organelles, and why these techniques are helpful in biochemistry
At about the same time that cell structure was being explored by microscopists, others were more focused on how cells functioned. What were chloroplasts and mitochondria doing, or the Golgi apparatus for that matter? What were chromosomes made of? How did cells stay alive? In the early 1800s, it was widely believed that living organism had some unique “vital essence” and were not subject to the laws of physics and chemistry. Biochemistry changed that by helping us understand what cells are made of and how they function. In this chapter, we will explore some of the key biochemical discoveries and techniques that helped move cell biology forward.
Enzymes and Biochemical Pathways
At around the same time that Schleiden and Schwann were articulating Cell Theory (Ch 1.1) in the early 1800s, a German chemist named Friedrich Wöhler demonstrated that the biological compound urea (found in urine of humans and many other animals) could be synthesized in the laboratory. This showed that a biological compound could be made from inorganic starting material, contradicting the idea that the “vital essence” of living organisms was somehow unique. This was an important paradigm shift for beginning to understand how the cells within organisms function, because much of that function (we now know) relies on chemical reactions. The field of biochemistry devotes itself to studying chemical reactions (the formation and breakdown of chemicals) in biological systems.
It took about another century to start understanding the importance of enzymes – proteins that help these chemical reactions occur in cells. Much of the early work to discover enzymes was done in yeast – single-celled fungal cells. In the late 1950s, Louis Pasteur showed that living yeast cells converted sugar into ethanol – a fact we still use to this day when making beer and wine (Figure 2.9). Another 40 years later, Eduard and Hans Buchner showed that fermentation could take place after yeast cells were broken open, but their contents were still active. The active agents from these yeast cells eventually became known as enzymes (“zyme” = yeast in Greek). We will learn more about enzymes in Chapter 6.4
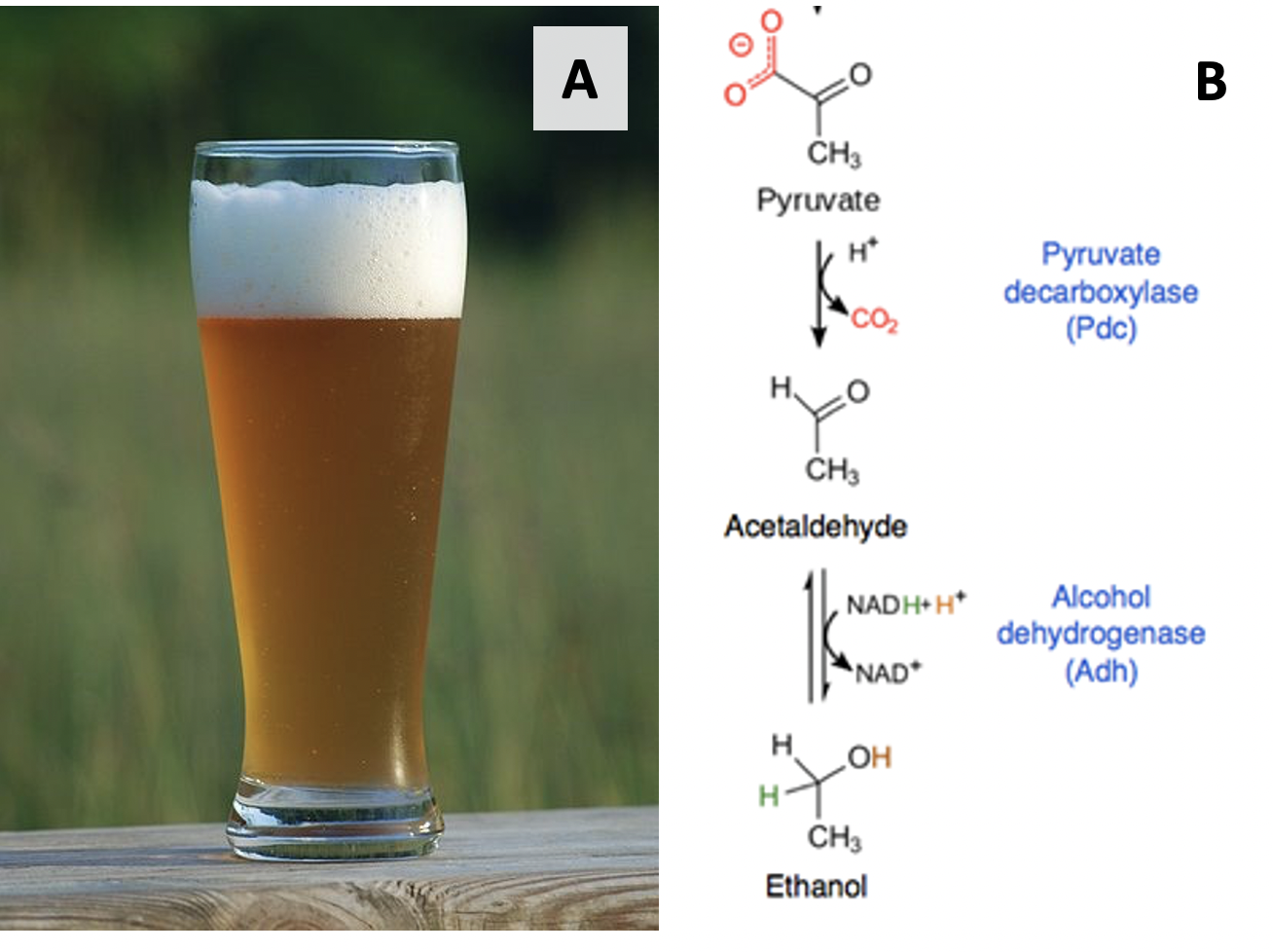
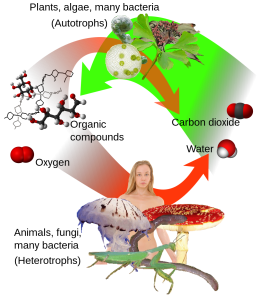
Most chemical transformations in cells require multiple steps. The example of alcohol fermentation in Figure 2.9 is a relatively short and simple pathway. In the early- to mid-1900s, various scientists started working out the individual steps in long and complex biochemical pathways. In the 1920s and 1930s, work by Fritz Lipmann showed that adenosine triphosphate (ATP) was an important molecular for energy transfers within cells. A team of biochemists – Gustav Embden, Otto Meyerhof, Otto Warburg, and Hans Kreb – described the multistep processes involved in breaking down glucose (and other molecules) to make ATP via glycolysis and the Krebs cycle (also called the tricarboxylic acid, or TCA, cycle), two pathways in cellular respiration. The Krebs cycle takes place in mitochondria, so characterizing this biochemical pathway helped improve our understanding of the role of mitochondria in metabolism. In the 1940s and 1950s, Melvin Calvin and his colleagues made an important step towards understanding photosynthesis by elucidating the steps of the Calvin cycle (also called the light-independent reactions) in algal cells. We now know photosynthesis is a core function of chloroplasts, allowing plants, algae, and their relatives to make sugars using energy from sunlight. These organisms then break down these sugars using cellular respiration and their mitochondria (Figure 2.10)!
Links to Learning
An overview of ATP & Cellular Respiration and Photosynthesis from Crash Course Biology.
Isolation of Cellular Components
To truly link function (e.g., metabolic processes) to particular organelles, it’s often necessary to actually isolate those organelles. Indeed, a lot of biochemistry requires purifying certain molecules or components so they can be studied in isolation. Many of the major developments in isolation techniques described below were important for characterizing the function and composition of organelles.
Centrifugation
How do you isolate mitochondria or chloroplasts or other organelles? One useful tool is centrifugation. Centrifugation is a way to separate and isolate substances based on their size, shape, and/or density. If we think about mitochondria from an animal cell, they somewhat intermediate in size – smaller than a nucleus, but larger than ribosomes. One can use a centrifuge (Figure 2.11a) to perform subcellular fractionation, the separation of cellular components. To do this, first you would break open your cells of interest (using something that damages cell membranes) in a liquid, and then put your liquid + cells samples in a centrifuge tube. When you operate the centrifuge it will spin rapidly, causing larger/denser components to move to the bottom of each centrifuge tube faster than smaller/less dense components. When material collects in the bottom of a tube, usually you end up with a solid pellet of that material, overlaid with the remaining liquid called supernatant (Figure 2.11b). By running the centrifuge at different speeds, you can control which components end up in the pellet or supernatant, and then work with your fraction of choice to better characterize its function.
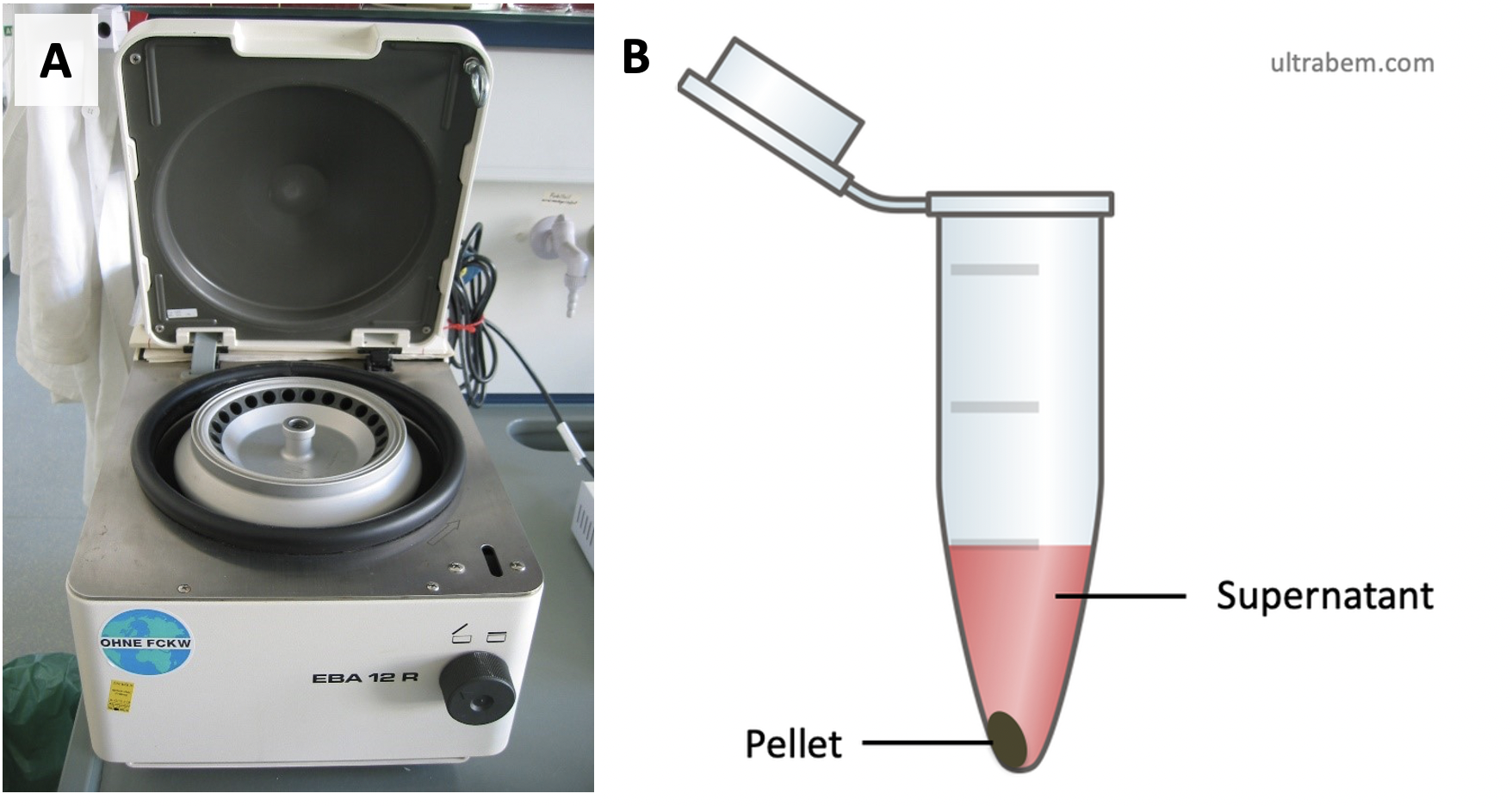
Ultracentrifuges are specialized centrifuges that are usually much larger than the benchtop one in Figure 2.11, which makes them well-suited for isolating very small organelles (e.g., ribosomes) and macromolecules (e.g., proteins). The ultracentrifuge was development by Theodor Svedberg in the late 1920s. You may have heard of 80S ribosomes and 70S ribosomes in eukaryotes and prokaryotes, respectively. The “S” stands for “Svedberg” (yes, the same Svedberg) and is measure of how fast a substance settles to the bottom of a solution under acceleration (e.g., during centrifugation). Ultracentrifuges can spin fast enough (e.g., 100,000 rotations per minute) to create forces that are more tha 500,000 times the force due to gravity. It is this force (usually expressed as a number ×g) that moves samples towards the bottom of a centrifuge tube.
Chromatography
How do you figure out which molecules are in an organelle? One technique that can help with that is chromatography. Chromatography includes a variety of techniques, all of which can used to separate a mixture of molecules into individual components based on their size, charge, or affinity (attraction toward) certain substances. The most familiar version of this is paper chromatography (Figure 2.12). For example, if you put a mixture of pigments (colourful molecules) at the base of a piece of filter paper, and then dip the base of some filter paper into that solvent, the mixture of molecules will start moving up the filter paper and separate based on their different chemical properties. In this technique, the paper is the stationary phase (does not move), and the solvent is the mobile phase (moving phase).
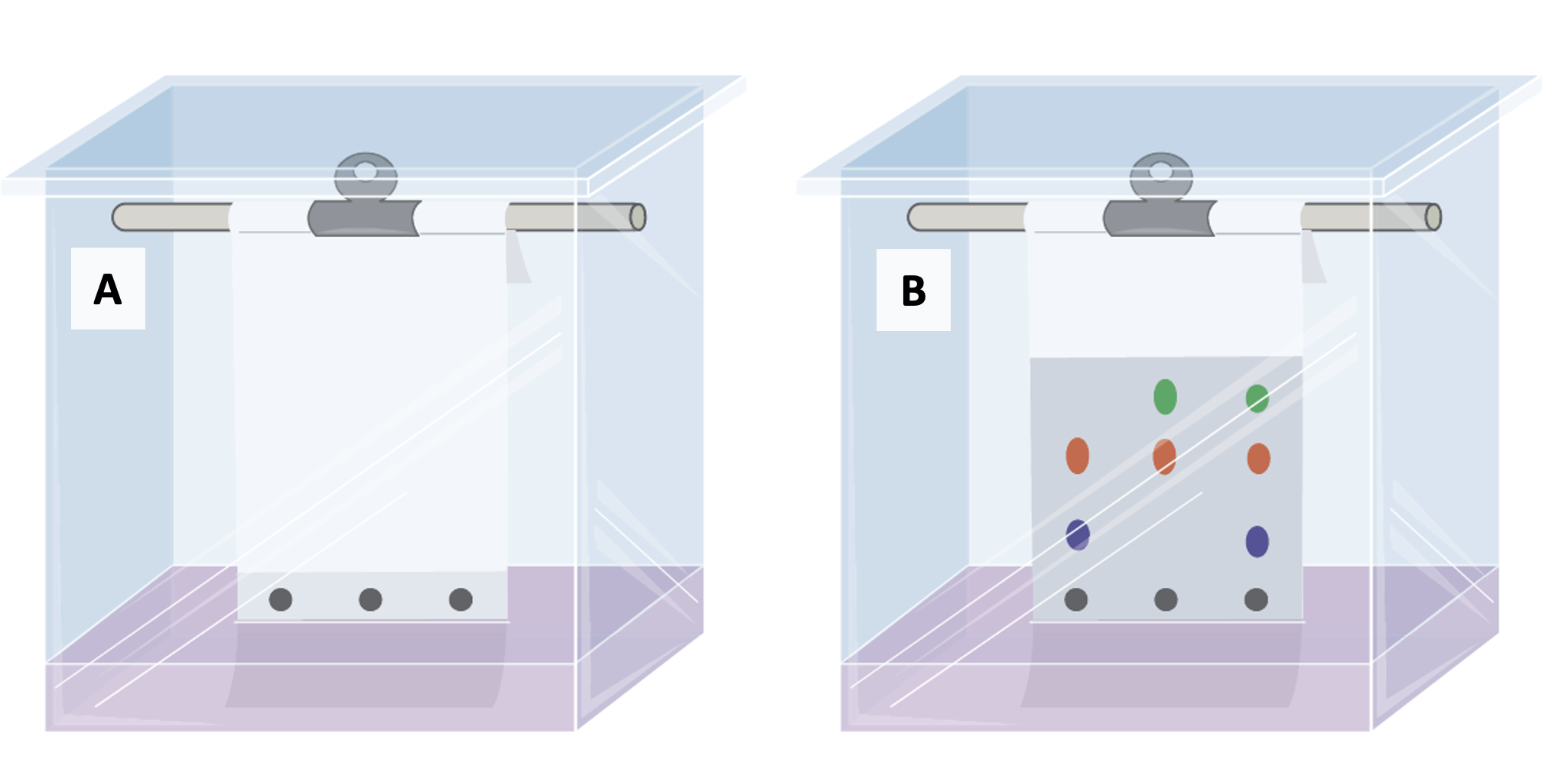
There are several other types of chromatography. Thin layer chromatography (TLC) has a similar setup to paper chromatography, but uses a thin layer of silica gel, alumina, or cellulose as its stationary phase. TLC can be a great way to test sample purity (one spot = one type of molecule in the sample). Samples do not have to be naturally pigmented (have their own colour) as long as the researcher can stain/visualize the spots in some way. Gas chromatography (GC) uses gas (rather than liquid) for its mobile phase, and the gas mixture moves through a column to facilitate separation of different molecules. Gas chromatography is sometimes coupled with a process called mass spectrometry (GC-MS), which is used to identify the molecules (chemicals) that were separated by GC. Gas chromatography can also be coupled with a flame ionization detector (GC-FID) to identify molecules, which is particularly useful for identifying lipids. Liquid chromatography (LC) is similar to gas chromatography, but uses a liquid mobile phase. Sometimes liquid chromatography is performed with the liquid under high pressure; we then call this high performance liquid chromatography (HPLC), and it can also be used to identify the molecules in the liquid phase. There are many other types of chromatography, but the last one we’ll mention is affinity chromatography. Affinity chromatography can be used to purify very specific molecules from a mixture based on the affinity between the chromatography column and the molecule of interest. It is often used in biochemistry to purify specific proteins.
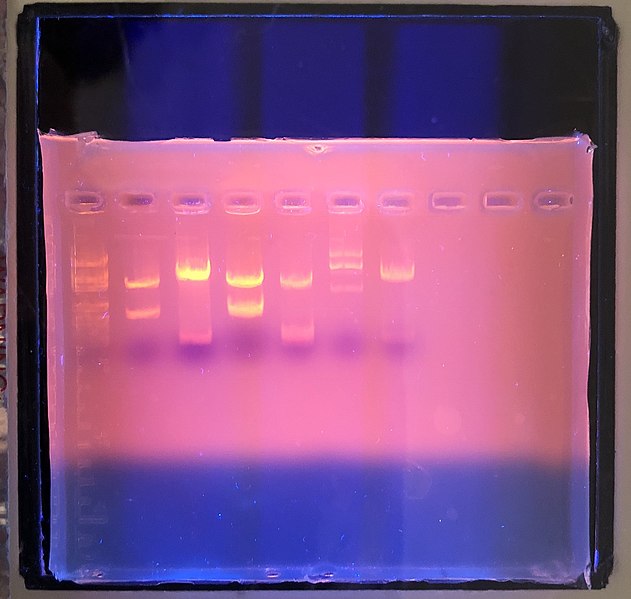
Electrophoresis
As we’ll discuss in the next section, genetics is also an integral component of cell biology. If you want to study DNA, RNA, or proteins, you’ll almost certainly end up doing electrophoresis. Electrophoresis includes a variety of techniques that use an electrical current to separate molecules based on their size and charge. Usually these molecules are moving through a semisolid gel, either made of agarose or polyacrylamide. Smaller molecules move faster through this gel, which larger molecules tend to move more slowly. This technique can be used to separate whole macromolecules (e.g., whole proteins), or fractions of macromolecules (e.g., segments of DNA or RNA). Once these macromolecules are separated by size, they can be isolated (e.g., cut out of the gel) and characterized, for example by figuring out the sequence of a particular protein or DNA molecule.