6 Chapter 6: Methods of Neuroscience
Austin Lim, PhD (DePaul University)
Claire Sun (Mount Sinai)
Editor: Katie L. Willis, Ph.D. (University of Oklahoma)
Most of the information you learn in neuroscience textbooks was discovered by people who applied the scientific method to systematically answer a research question. In order to come to their conclusions, these neuroscientists used a variety of techniques, borrowed from biology, medicine, chemistry, physics, engineering, psychology, and many other academic disciplines. Neuroscience researchers use the wide range of scientific methodological strategies to answer: “How does the nervous system work?”
In this chapter, we will describe a few of the methods that are used to ask and answer questions in neuroscience. These methods can be divided roughly into four major categories: Imaging anatomy, imaging function, imaging cells, and finally manipulating the nervous system. Within each of these categories, the techniques will start with the “big picture” view, and by the end of each section we zoom down to the level of molecules and genes.

Just a minor caveat about these techniques. Many of these strategies can be used independently, but some may be used simultaneously.
There are three major components that can be described for some of the techniques:
- A description of how the method works. Each of these methodological strategies rely on using the knowledge from other fields of study and applying that knowledge to a question about the nervous system. Knowing how the methods work allows us to decide what types of data can be collected and when each method will be appropriate to use.
- Some of the main advantages of the technique. Not every method is applicable for studying every question. Instead, it would be beneficial for neuroscientists to choose the most appropriate strategy for a particular research question given the circumstances.
- Some shortcomings or limitations of the technique. As above, no strategy is perfect. Methods that look at the “big picture” aren’t able to look at microscopic level detail. There are limitations for every technique and being aware of these limitations inform us about caveats in interpreting data.
About resolution
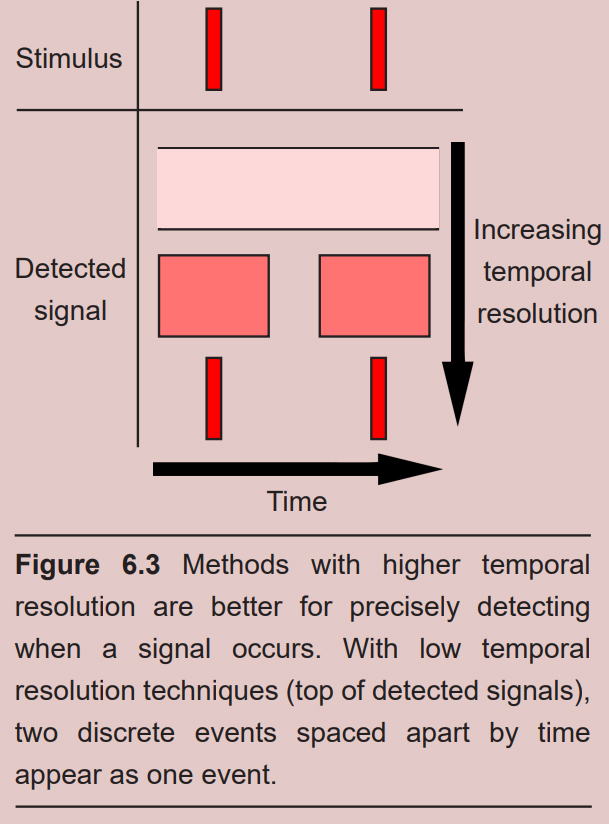
Many of the imaging techniques are concerned with resolution. Resolution in neuroscience is very similar to resolution in digital photography or computer monitors. A picture taken in high resolution contains more information than one in low resolution and can, therefore, be more valuable. Imaging strategies consider two different types of resolution, spatial and temporal.
Spatial resolution refers to the ability to differentiate two points in space from each other. An imaging method with high spatial resolution means that two signals very close together can be identified as two different signals instead of one. Spatial resolution is usually measured in units of distance or volume. Electron microscopy (described in 6.3.1) offers the highest spatial resolution of the imaging techniques.

Temporal resolution refers to the ability to distinguish two events in time from one another. Temporal resolution is measured in units of time. Functional imaging techniques with high temporal resolution, such as electrophysiology, can differentiate two signals as close together in the hundreds of microseconds range.
Although almost all these techniques can be used in either humans or non-humans, some techniques are best adapted for humans and others for non-humans. Using a technique on an inappropriate species can lead to poor results. For example, humans can lay still for tens of minutes at a time during an imaging scan session, while rats or mice will not be able to do this simple behavior (without restraint or anesthesia). Before getting started with our list of research methods, we should ask ourselves “Is it better to study the question in humans or in non-human animals?”
The case for human subjects
The main reason we conduct studies in humans is to understand the human nervous system. It is knowledge of the human condition that leads to the development of therapies that prevent, cure, or otherwise improve the quality of patient’s lives. And while curing diseases in non-human animals is the purpose of veterinary medicine, curing humans is the primary end goal of biomedical research, and that goal is only truly met by testing on humans. A cure developed for rodents might give us a clue about the human case, but without seeing how well it works in humans, the therapy won’t save human lives.
Humans are great at following directions without much training, unlike non-humans – as anyone who has tried to house train a puppy can attest. We can follow simple sets of directions that untrained non-humans find difficult, such as “lie still”. During functional activity scans, we can perform extremely complex higher-order cognitive tasks, such as “imagine that we give you 20 dollars, and we will take away a dollar when you answer a question incorrectly” (rats and chimpanzees have no concept of currency). We can describe our feelings in writing and self-report on our symptoms, both of which are tasks that are impossible in non-humans.
Believe it or not, recruiting human subjects for neuroscience or psychology studies is usually cheaper than studying that same question in nonhuman animal models. Collecting data from undergraduates is sometimes free, since many are required to participate in some number of hours of psychology studies to pass introductory level classes. Although the cost per patient differs depending on the nature of the study, many patients get paid as little as $10 an hour. On the other hand, housing colonies of mice, rats, or monkeys gets very expensive, very quickly.
The case for non-human subjects
But, it is often impossible to test every question in humans. Instead of always studying humans, scientists often use non-human model organisms, the most common organisms being the worm C. elegans, fruit flies (Drosophila melanogaster), zebrafish (Danio rerio), song birds, mice, rats, and macaque monkeys. The closer we move towards the human, the more similarities the model organism shares with us. Of the commonly used model organisms, macaque monkeys are the non-humans that are most similar to humans. We share 93% of our genetic material with macaques, but we still have different metabolic and physiological processes, and our behaviors are much different from theirs.

Ethical constraints prevent us from performing experiments that may cause physical or psychological harm if performed in humans. We would never conduct a test on humans to assess what concentration of neurotoxin leads to brain damage (these experiments aren’t done very frequently in nonhumans anyway.) Invertebrates, such as worms and fruit flies are not heavily regulated by ethics oversight committees, allowing scientists to conduct a wider set of experiments on these animals.
Experimental preparations
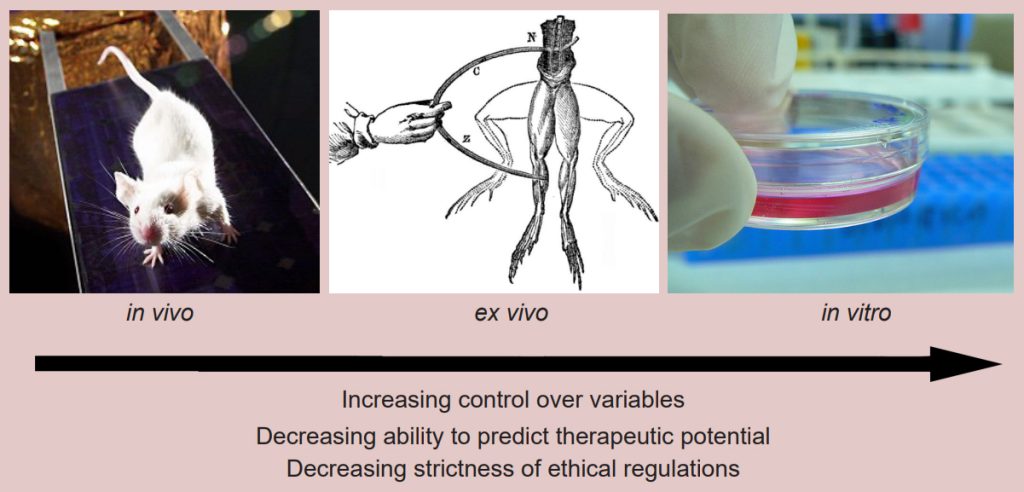
Performing an experiment in an intact, living organism, whether human or nonhuman, is described as an in vivo (Latin meaning “within life”) preparation. The main strength of this strategy is that the data collected here are more predictive of the human condition, which is one of the main goals of biomedical research. However, the in vivo preparation has challenges, because thousands of variables within a living system are uncontrolled or still unknown. There are also very strict ethical limitations on the nature of experiments that can be done in vivo.
On the other hand, an in vitro (Latin meaning “within glass”) preparation is an experiment performed on cultured cells or isolated molecules of DNA, RNA, or protein. These preparations have the opposite strengths and weaknesses of in vivo preparations. They allow for extremely good control over variables, but the results are less reliable in translating to a therapy. The regulations on these experiments are much more lax compared to in vivo experiments; most of the regulatory guidelines are to protect the experimenter rather than the patient or the experimental subject.
Falling in between these two preparations is an ex vivo experiment. In this kind of experiment, a section of the living organism is taken, such as a slice of brain, a tissue biopsy, or a detached frog leg. The strengths and limitations of these experiments are somewhere in between that of the other two preparations.
Nonhuman animals perform all kinds of behaviors that humans never experience. These behaviors are unique to nonhumans and can only be studied in nonhumans of course. We will never be able to understand flying (birds) or slithering (snakes) as a means of locomotion by studying humans.
One thing that makes human studies difficult is how weird we are. We all come from very different backgrounds, bringing our own set of experiences, flaws, and biases into every research study. Your current mood and mind state can alter behaviors profoundly – just ask any one who gets “hangry.” In nonhuman studies, many of these variables are closely monitored. When we raise model organisms in the lab, almost every aspect of their life is controlled, from their food source, living conditions, and their day- night cycles, which eliminates several sources of variability that affect human studies.
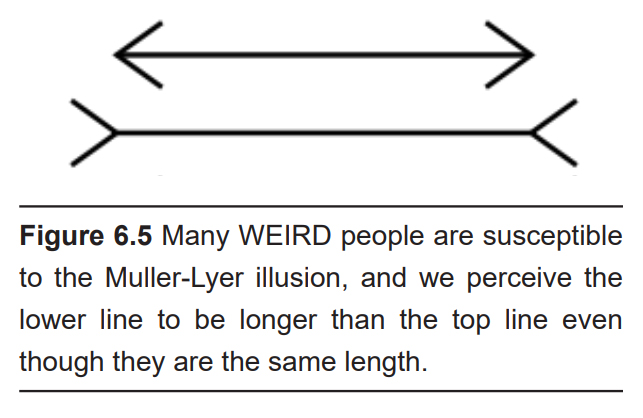
Another factor that makes human studies challenging is that we are WEIRD: The acronym meaning that most patients in psychology studies are Western, Educated, and from Industrialized, Rich, and Democratic countries. One study estimates that about 96% of psychology studies use WEIRD subjects, whereas these people are only 12% of the total global population. This creates a bias in sample selection, since WEIRD people perform differently on behavioral tasks compared to others. Even our susceptibility to visual illusions is affected by our WEIRD-ness.
Chapter 6 outline
6.1 Imaging brain activity
6.2 Imaging brain function
6.3 Imaging the cells of the nervous system
6.4 Changing nervous system activity
6.1 Imaging brain activity
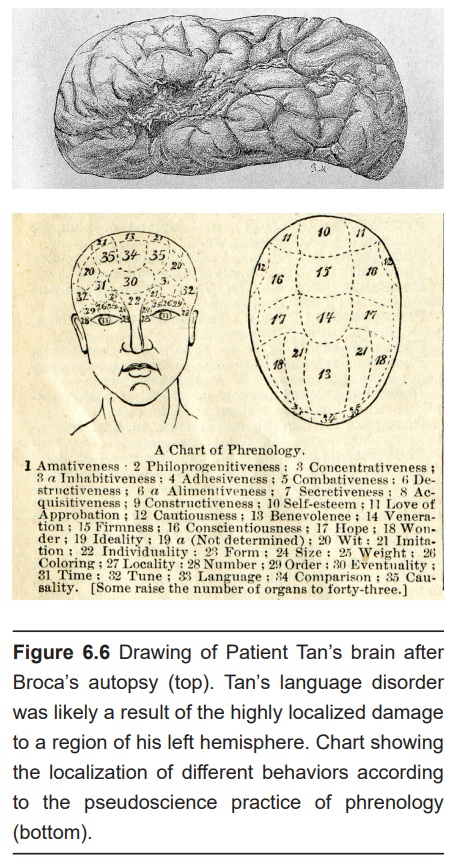
The earliest methods of analyzing nervous system anatomy were crude: manual dissection of the brain post-mortem (after death). In the 1860s, these methods received attention clinically with the studies conducted by French neurologist and surgeon Paul Broca. One of Broca’s patients, nicknamed “Patient Tan,” had severe language deficits and was only able to say “tan.” Broca performed an autopsy after Patient Tan died and observed a very localized injury in his brain that was likely the cause of Tan’s language deficits.
At the same time, a pseudoscientific fad was gaining in popularity. Franz Gall, a German neuroanatomist, published a treatise describing phrenology, the belief that we can predict personality traits based on the shape of a person’s head and the bumps on the outside of the skull. According to Gall, behaviors such as romanticism, individuality, and cautiousness are a result of differences in brain anatomy that push the skull outwards.
Today, we know that phrenology is a fad unsupported by the rigorous methodology of science. But there was a valuable lesson that emerged from phrenology which still persists in part today. Both phrenology and Broca’s observation support localization of function, the idea that specific areas of the brain are important for certain functions. Today, we also think about the connections between two areas as being important for healthy brain activity. The techniques discussed below were developed to allow scientists to see some aspect of the anatomy of the nervous system, either gross anatomical differences or connectivity.
6.1.1 Computerized tomography scan (CT scan or CAT scan)
Example questions answered:
“Does the patient have a brain tumor, and where is the brain tumor located?”
“Are the meninges intact?”
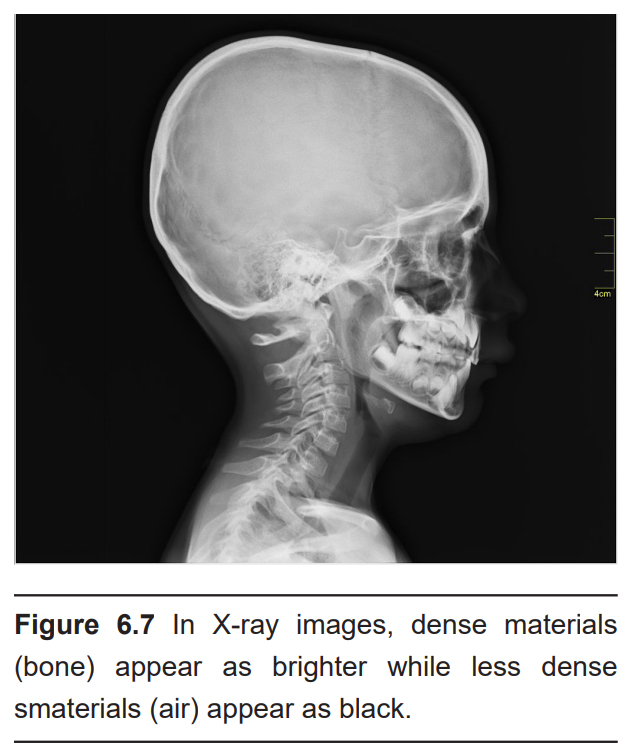
The CT scan relies on X-ray technology that was developed in 1895. X-rays are high energy beams of electromagnetic radiation that are capable of passing through many physical objects. Traditional two-dimensional X-rays, such as those used to image a broken bone or tooth decay, use radiographic film to detect where the X-rays get blocked. When an X-ray passes unimpeded, it causes the film to darken. But, wherever the X-rays are blocked, the film remains white. Therefore, material that is more dense (such as bone) appears as white, less dense material (such as the air surrounding the body or CSF) appears dark. Other tissue are some shade of gray in between.
(Can we fix the caption to spell materials correctly?)
The CT scan is essentially a three- dimensional X-ray that revolves around the person as they move through the scanner. Instead of using radiographic film, the CT scan uses a computer that detects the passage of X-rays, located directly across the emission source of the X-ray. Instead of a flat, two dimensional-image, the CT scan uses an X-ray gun that revolves around the person’s body as they advance slowly through a large circular hole. The computer is then able to compile the series of two-dimensional images and turn them into a three-dimensional reconstruction that can be used to see the brain from any projection. CT scans give us a spatial resolution of about 0.5 mm. CT scans are generally used clinically to assess diagnostic changes over several days (such as before and after tumor removal or to determine if an intracranial bleed has healed), so temporal resolution is not a major consideration.
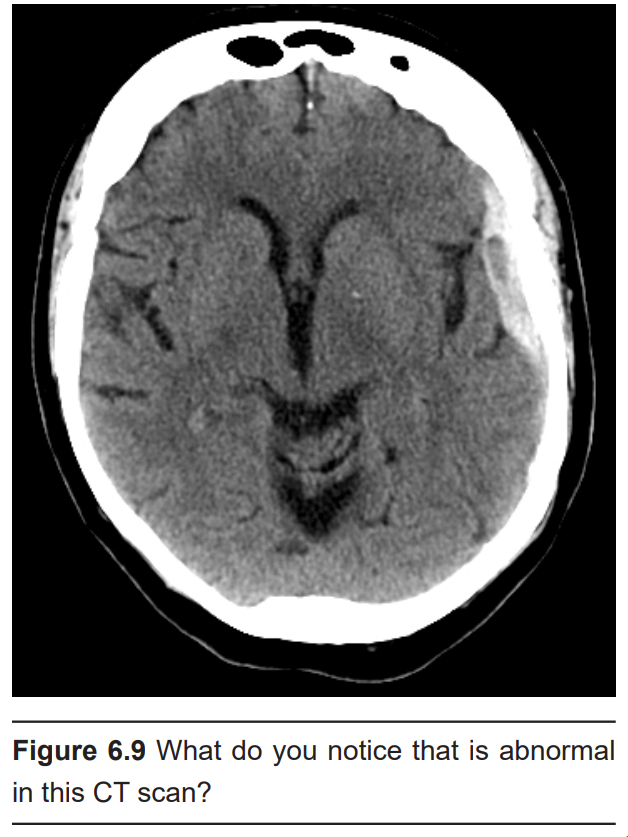
As an anatomical analysis that can easily identify tissues of different density, it is great for identifying and diagnosing particular brain conditions. Brain tumors can be visualized in a CT scan, since they are identified by an increase in tissue density compared to normal brain tissue. Hydrocephalus, an abnormal and potentially deadly expansion of the CSF-filled ventricles, can be quickly identified by this analysis. Meningitis, an inflammation of the meninges, may present as increased contrast in the CT scan.
The big advantage of the CT scan is that it is noninvasive. You can use a CT scan in order to diagnose and identify the cause of a condition while a person is still alive and, hopefully, work towards developing an intervention.
It is also a relatively quick technique. A full head CT scan takes only minutes, which allows for a rapid diagnosis of anatomical structures.
However, X-rays are highly mutagenic. Prolonged exposure to X-rays dramatically increases the risk of developing various cancers, since X-rays interfere with the process of DNA replication. It is estimated that the radiation exposure in a single head CT scan is similar to the background exposure of X-rays in a few months. When a CT scan is prescribed, the diagnostic information gained from a CT scan is more important than the risks from increased radiation exposure.
6.1.2 Diffusion tensor imaging (DTI)
Example questions answered:
“Is the volume of the white matter tract medial longitudinal fasciculus important for normal language processing?”
“Does spinal cord compression cause neurological deficits?”
While a CT scan is great for detecting gross anatomical anomalies like tumors or intracranial bleeding, it has a difficult time with subtle anatomical changes like differences between gray matter and white matter tracts. A technique for identifying these differences was proposed in 1994, called diffusion tensor imaging (DTI).
DTI quantifies white matter because of the morphological features of white matter. More specifically, DTI uses MRI technology (see section 6.2.3 for more information) to detect and quantify the movement of water molecules, which move differently in gray matter than white matter. A single molecule of water in the middle of a cup can move in any direction with equal probability. This type of motion is called isotropic diffusion. However, the movement of a water molecule in biological tissue is not completely random, largely because the brain is made up of heterogeneous tissue. White matter is very different from gray matter. A water molecule is more easily able to diffuse along the same direction as a tract of white matter, but has a difficult time moving perpendicularly across such tissue. The difference in molecular motion is called anisotropic diffusion. DTI uses the premise that anisotropic diffusion is observed in white matter tracts.
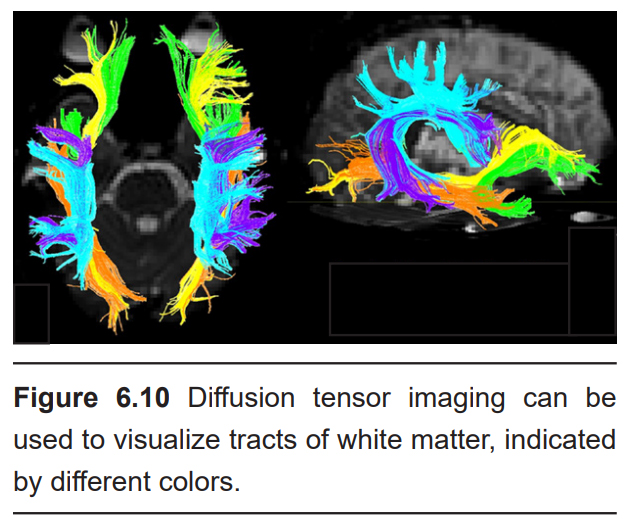
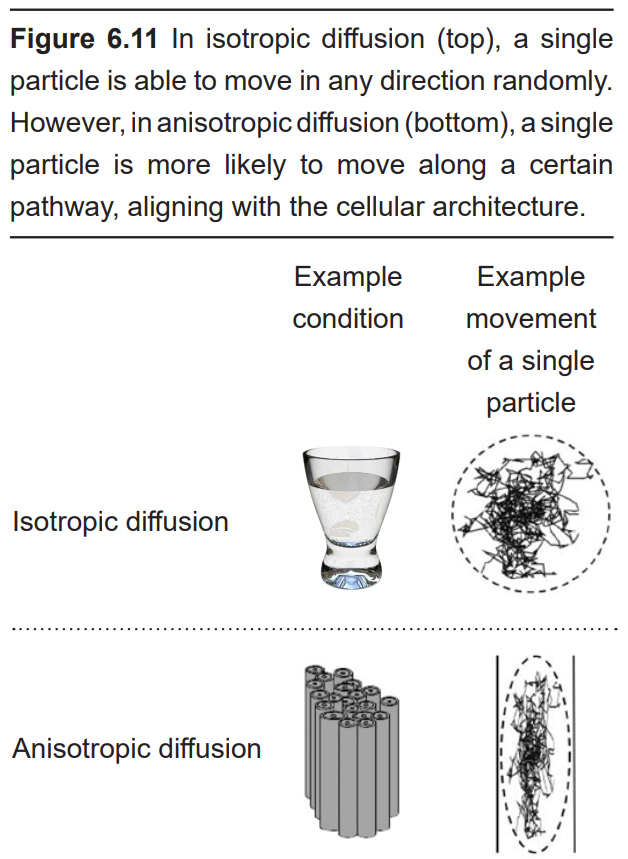
DTI can give spatial resolution on the order of millimeters. It is generally not used for monitoring changes over time, so temporal resolution is not a major consideration.
Axonal projections are directional, with the soma at one end and the axon terminal at the other. One of the shortcomings of DTI is that it cannot give us information about the directionality of the axonal projections.
6.1.3 CLARITY
Example questions answered:
“Do neurons in layer 5 of motor cortex send axonal projections into the spinal cord?”
The methods described above only provide rough anatomical information, and therefore have limits to their value. CLARITY is a revolutionary anatomical technique that gives microscopic level analysis. First published in 2013, CLARITY was developed at Stanford by the lab of Dr. Karl Deisseroth. The value behind CLARITY is that we are now able to visualize connectivity in the brain at a microscopic level, giving us amazing spatial resolution at the order of a microns.
One of the problems with visualizing the structures of the brain at a microscopic scale is that cells have a lot of membrane, which is mostly made up of lipids. Instead of letting light pass unimpeded, lipids refract light in somewhat unpredictable ways. Even cytosol has all sorts of particles floating around in it that cause light to bend and bounce around. These lipids and other cellular particles prevent light from passing through nervous tissue, minimizing our ability to use transmitted light to see the brain.
In CLARITY, the brain is first flushed with chemicals that form a gel matrix that surrounds every cellular structural component, from the spines on the dendritic arbor all the way to the axonal terminals. Then, using a chemical detergent, the cellular lipids get solubilized and washed away while the gel matrix remains unaffected. This allows us to now see the “mold” that remains around where the cell membranes used to be. By washing away the light-deflecting lipids, we can now see where the connections were, causing the brain to appear transparent.
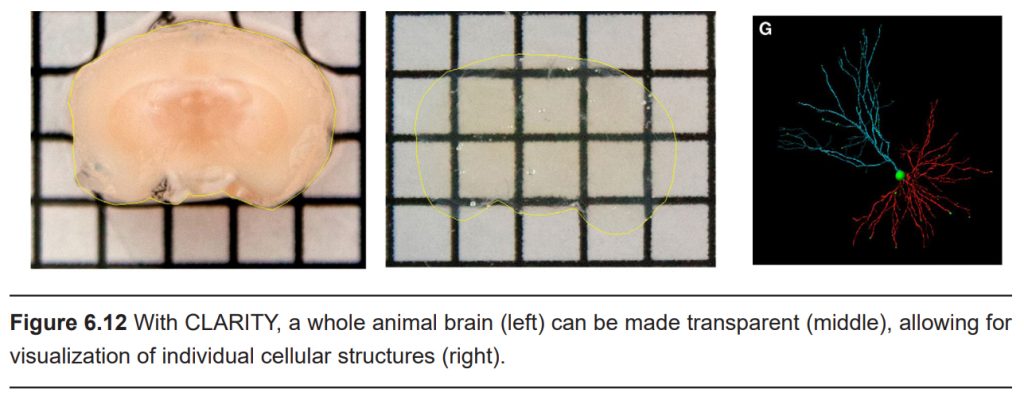
Unlike the CT scan, CLARITY can’t be applied in an intact living organism. CLARITY is an extremely destructive process that destroys tissue. The surrounding gel matrix is like a mold of a hand. You can see all the anatomical features of the hands – fingers, wrinkles, nails, and so on. But the mold is just an image, unable to function. The mold of the hand cannot grip things, cannot feel things, and does not have any bones / tendons / muscles. In using CLARITY, all function is destroyed.
6.2 Imaging brain function
Anatomical analyses are certainly a meaningful way to study aspects of the nervous system. But being able to measure and record brain function is important for answering different types of questions. After all, the brain of a recently deceased person might look identical anatomically to that of a living person, but they would exhibit significantly less function.
6.2.1 Electroencephalography (EEG)
Example questions answered:
“At what times of the night does the sleeping brain exhibit synchronized neuronal activity?”
“How does brain activity change when a person is having a seizure?”
The electroencephalogram (EEG) is one method by which we can observe electrical activity of the brain. When neurons send action potentials, several charged ions move across cell membranes, which causes a change in the electrical charge of the adjacent area. Some of these charges are very large, especially when several millions of neurons are sending action potentials at the same time. These currents, produced by neurons of the outer surface of the brain (cortex), can be so large that they are detectable outside the skull at the surface of the head. The EEG works by recording these changes at different areas of the brain. The technique was first used in the 1920s by psychiatrist Hans Berger, who invented the technique. The theory behind the EEG is the same used in an electrocardiogram, or ECG, to detect the electrical activity of heart cells.
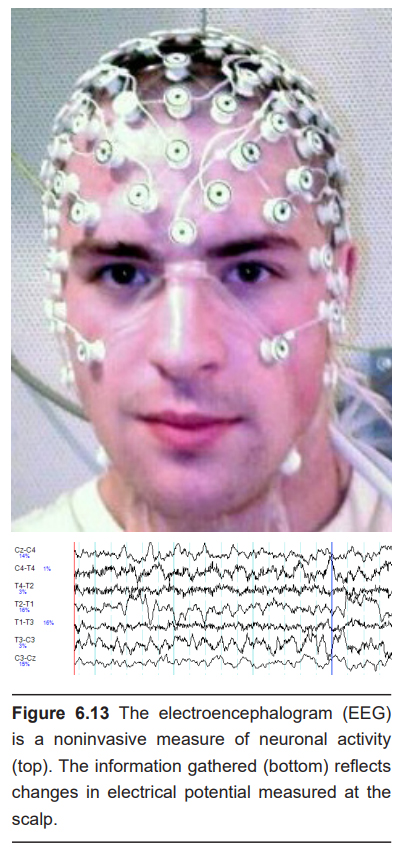
To perform an EEG, a gel is applied onto small patches of the scalp. This gel is basically a sodium chloride gel that acts as a conductor, allowing the currents to be picked up by a series of adhesive electrodes. EEG systems used in hospitals for diagnosis or in laboratories for research studies may have anywhere between 20 to 128 electrodes. Each electrode is sensitive enough to detect voltage deflections as small as 10 microvolts. Each electrode is placed on a specific area of the scalp, and the information from each electrode runs into a computer. From there, the computer can then perform calculations to determine which cortical areas to the brain exhibit what patterns of activity.
EEG software is capable of examining the rapidly-fluctuating electrical potentials and dissecting out the several components buried within the complex signals. From one waveform, it can pick out high frequency components called beta waves (between 13 and 30 Hz), low frequency components called delta waves (between 0 and 4 Hz), and all frequencies in between.
The EEG is a noninvasive functional analysis method. Nothing permanent is done to the person when they get an EEG because it is only detecting information. Wearing a cap of electrodes may be annoying, but it is a harmless procedure. Because of the experimental set up for EEG, there are many cases where using an EEG is preferable. The EEG is a standard part of a polysomnogram, a series of tests that can be done to evaluate sleep disorders. In fact, our best and most reliable characterization of the phases of sleep rely on brain activity measures as recorded by an EEG (Chapter 13). Relatedly, EEGs are also used clinically when a person is under anesthesia as a tool to evaluate their level of unconsciousness.
Not only is the EEG noninvasive, but it is also relatively cheap – at least in comparison to many of the other methods described in this chapter. It does not require heavy equipment and is mobile. All the machinery needed to run an EEG can be contained within a backpack.
EEG is also useful for the diagnosis of a variety of brain disorders that are a result of aberrant cortical neural activity. The most well-known of these is epilepsy. In epilepsy, when a person has a seizure, it is believed that large masses of neurons are sending action potentials across the cortex, producing very large signals that can be detected by scalp electrodes. Migraine, a debilitating condition, may be due to waves of unusual neural activity that sweep across the cortex. Other applications of EEG may include helping diagnose Alzheimer’s disease, depression, and possibly ADHD.
The EEG has great temporal resolution. Because it is detecting changes in currents, it is capable of sampling in the range of 10,000 Hz, meaning that for each electrode, 10,000 data points can be collected per second. Since action potentials happen on the time scale of milliseconds, being able to calculate changes in electrical potentials at such speeds gives us the ability to assess brain activity very precisely.
But, the EEG has poor spatial resolution, only detecting signals that originate in the outer most layer of the brain. More electrodes increase spatial resolution, but even at 128 electrodes, an EEG only has spatial resolution to the level of about 7 cubic centimeters.
6.2.2 Positron emission tomography (PET scan)
Example questions answered:
“Which areas of the brain decrease in activity when a person experiences mild cognitive impairment?”
“Do drug-dependent people have a high density of opioid receptors?”
The positron emission tomography (PET) scan is an application of nuclear medicine best known for its applications in the medical setting for the diagnosis of cancer. Before the PET scan, a radioactive compound called a tracer is injected into the bloodstream. This tracer is a compound where an atom is substituted with a radioactive isotope, such as tritiated hydrogen (3H) or fluorine-18. The PET scanner itself is a large circular device that looks similar to the CT scanner. The tracer is chemically unstable, and it spontaneously emits positrons. When those positrons interact with the electrons of nearby molecules, two gamma particles are emitted in perpendicular directions. These gamma particles are then detected by the PET scanner as the person moves through the machine.
A common tracer that is used for imaging brain function is fluorodeoxyglucose-F18, or FDG. FDG is a radioactive analog of glucose, one of the main sources of cellular energy. Highly metabolically active areas of the body take FDG into the cells. Energetically demanding areas of the brain require more energy to do their functions (the same reasons that tumors, rapidly- reproducing patches of tissue, appear very bright in PET scan analyses). In order to produce the energy needed for increased neuronal activity, the brain changes perfusion levels by dilating blood vessels in order to bring more glucose to those areas. Therefore, when an area of the brain increases in energetic demand, that change can be detected by identifying the increase in glucose movement.
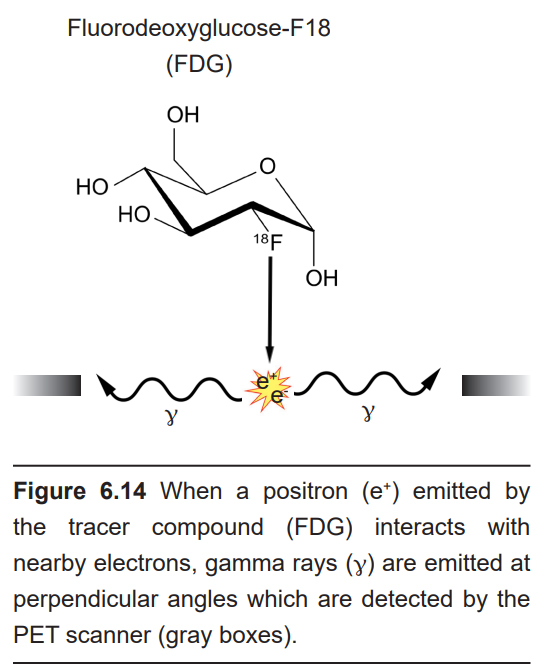
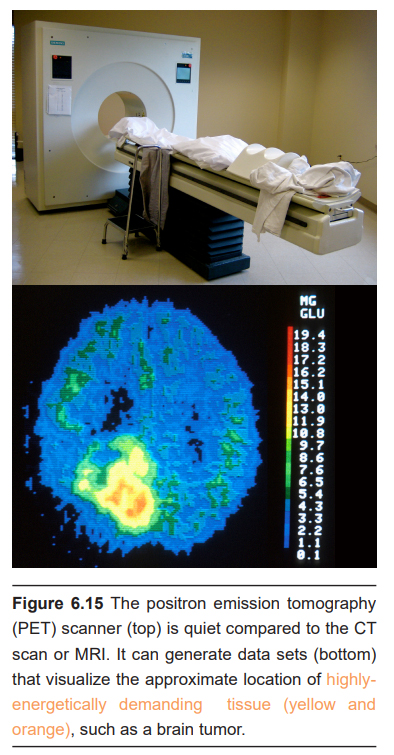
PET scans can be effective at diagnosing and identifying the location of tumors in the nervous system. It also provides an overall picture of brain activity, which may be useful in diagnosing disorders of cognitive deficits, like dementia associated with Alzheimer’s disease or Pick’s disease. PET scans have also been used to image the activity of specific brain areas as a person performs behavioral tasks, but this use of PET scan has largely been replaced by functional magnetic resonance imaging (fMRI; see section 6.2.3).
A second application of PET scanning is to visualize levels of receptors in vivo. This experimental approach relies on the nature of the interaction between specific radioactively-labeled compounds and receptors (such as [11C] raclopride and dopamine receptors, or [18F] ASEM and acetylcholine receptors). Once the radiolabeled compound binds to the receptor, the PET scan is able to detect the location of the radioactive signal and the strength of that signal. The downside of the PET scan as a diagnostic tool is similar to a limitation of the CT scan. A person is exposed to radioactive compounds and gamma wave radiation, which are potentially mutagenic.
In images produced by a PET scan, it is often difficult to identify boundaries between tissue, even between dramatically different internal organs. To make up for this deficit, PET scans are frequently performed simultaneously with an anatomical analysis like a CT scan.
PET scans generally have very poor spatial and temporal resolution. With PET scanning, you can really only see differences between areas if the volume is in the range of 5-10 cm3. Anything smaller than a cubic centimeter will be impossible to detect with current PET scanners. As to the speed of the PET signal, it often takes tens of seconds or minutes before a change in signal can be observed and detected.
6.2.3 Functional magnetic resonance imaging (fMRI)
Example questions answered:
“Do neurons in the right hemisphere cingulate gyrus increase in activity when a person sees their loved ones?”
“Which areas of the brain change in activity when a person is planning a motor action?”
The functional magnetic resonance imaging (fMRI) technique is probably the most well-known method of studying brain activity. Because fMRI can be performed while a person is engaged in a task, many research studies use fMRI as a means to correlate behavior with activity patterns in specific parts of the brain.
An fMRI machine basically consists of two main components. As with the CT scan or PET scan described above, the fMRI machine is a circular tunnel through which a person on a table moves. As the person moves through the scanner, an extremely powerful magnet revolves around their head. The power of a typical magnet used in a hospital fMRI may be as powerful as 10,000 gauss (1 Tesla), strong enough to lift a car. The more powerful fMRI machines can be as powerful as 100,000 gauss (10 Tesla). The stronger the magnets, the better the spatial resolution that the machine can produce (our current best spatial resolution is on the order of millimeters). The second component is a device that emits radio waves. Just like the magnet, the radio emission device revolves around the head.
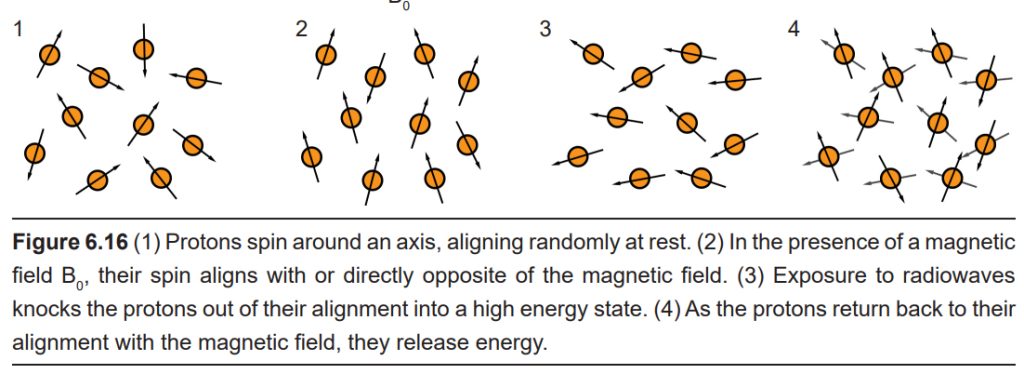
Both the magnetic field and the radio waves are crucial for the fMRI to work. They both interact with protons, the charged subatomic particles that have a small polarized direction because they spin around an axis. Initially, when these protons enter into a strong magnetic field, each proton charge aligns either with or directly against the direction of the field. Then, when hit by a radio wave, the protons lose their alignment, going into a high energy state. The protons then fall back to a low energy state as they return to alignment with the magnetic field. This process is what is being detected by the fMRI.
As it turns out, the protons in oxygenated hemoglobin are sensitive to the magnetic field (diamagnetic) while deoxygenated hemoglobin is not (paramagnetic). Because of this difference, we are able to use sensitivity to a magnetic field as a measure of changes in oxygenation levels. Like the PET scan, the fMRI hinges on the idea that more active areas of the brain have different metabolic demands than less active areas of the brain. When there is more activity in one area of the brain, the neurons in that area need more oxygen, and the adjacent blood vessels react by dilating. This change in blood flow is detected by the fMRI, and is called the blood oxygenation level-dependent signal, or BOLD signal. Temporal resolution of the fMRI is limited by the speed of blood vessel dilation, which occurs on the order of seconds to tens of seconds.
The main reason fMRI is useful in so many research applications is that you are able to visualize brain activity real-time during the performance of complex behavioral tasks. You can present specific visual stimuli to a person in an fMRI scan and evaluate which parts of the brain changes in activity. For example, seeing pictures of faces causes increased blood flow into the fusiform face area. You can ask a person to perform a gambling task and evaluate the areas of the prefrontal lobe that are responsive to risk taking.
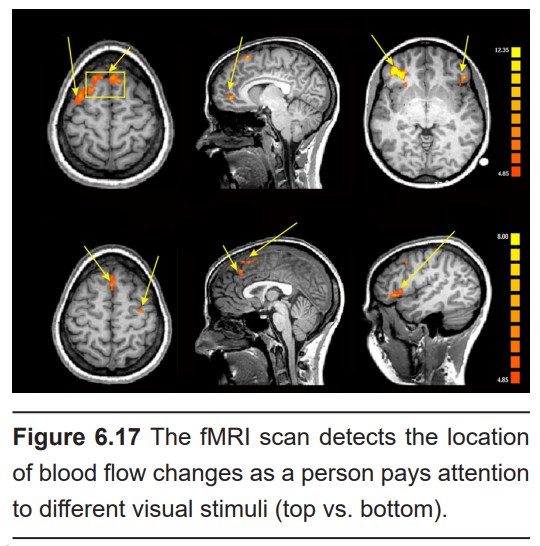
Although the technique has a great capacity for analyzing brain function, the nature of the machine itself presents limitations. The scanning tunnel is very small and claustrophobic, making it difficult to safely study anxiety or panic disorder without endangering the patient. The machine can also be very loud, which is not trivial if you are interested in studying younger patients. The use of a tremendously powerful magnetic field presents a different set of limitations. At the risk of severe injury or death, the patient entering the scanner cannot have any magnetosensitive implants, such as metallic aneurism clips, intrauterine devices, or shrapnel. Even older generation tattoos have trace amounts of metal that cause burns when exposed to the magnets of the fMRI machine.
The data collected by fMRI can be very difficult to analyze and are frequently subject to false positives. The change in BOLD signal measured in an fMRI is very small: one study estimates that perfusion increases by only 0.4% even under the greatest activation. Using standard fMRI analysis methods, an infamous study demonstrated that even a dead salmon in an fMRI exhibits brain activity that is similar to those reported in other fMRI studies.
FMRI also assumes that increased blood flow is directly correlated with the amount of neural activity, which may not always be the case.
6.3 Imaging the cells of the nervous system
The methods thus far discussed are limited in how closely we can see the components of the nervous system. For example, it could be useful to try and see the specific cellular features of a healthy neuron and compare that with the features of a neuron that has been damaged by multiple sclerosis. To be able to visualize the morphology of structures smaller than hundreds of microns, you’ll need a different set of techniques.
6.3.1 Microscopy
Example questions answered:
“What is the diameter of the soma of a granule cell?”
“How are the retinal neurons at the fovea different from the neurons in the periphery?”
Among the techniques described in this chapter, short of physical dissection or autopsy, microscopy may be the oldest. Microscopy began with the Dutch scientist Antonie van Leeweunhoek in the late 1700s. Originally, microscopy was more of a novelty for entertainment rather than a piece of equipment fit for a laboratory. Using a series of precisely-crafted lenses and a bright source of light, Leeweunhoek was able to see tiny animals living in water.
Microscopy gives us the ability to see things at a level of resolution that would otherwise not be possible. Compared to today’s microscopes, the earliest scopes provided poor magnification, able to bring us only about 40 times closer than the unaided eye (these days, a standard laboratory microscope provides a level of magnification up to 400 or 1,000 times closer). Other types of microscopes, such as the electron microscopes developed in the 1930s, use an electron emission device in conjunction with high speed detectors to visualize structures that are on the order of nanometers, giving us a zoom of up to a million times!
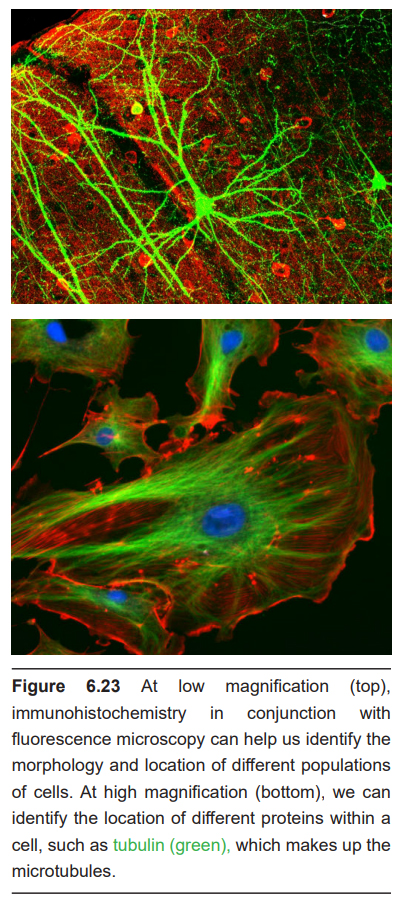
A different form of microscopy relies on the fluorescent properties of molecules such as green fluorescent protein (GFP). Fluorescence microscopes use light sources such as filtered light or lasers which emit light at specific wavelengths. Photons at specific wavelengths activate the proteins, which in turn emit light at a different wavelength. The emission of this second color of light can then be detected.
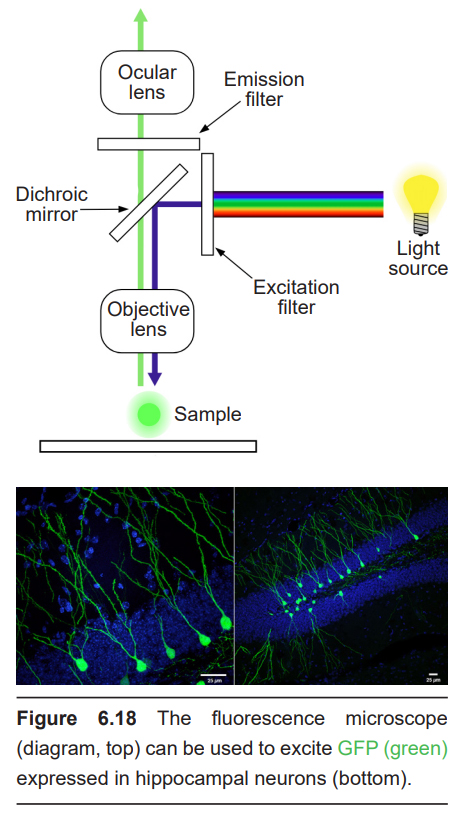
Microscopy is a standard part of a neuroscience research kit. Most of the techniques below will rely on microscopy to visualize some component of the nervous system.
6.3.2 Staining
Example questions answered:
“Where is the white matter located in the brain stem?”
“What is the morphology of dendritic arbor of a cerebellar Purkinje cell?”
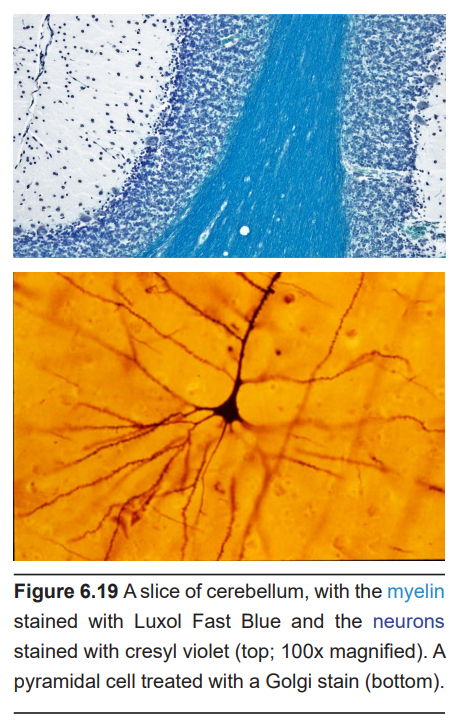
Staining is an imaging method that is often used in conjunction with microscopy. Thin slices of brain tissue are exposed to various chemical processes. The chemicals that are used for staining have different affinities for parts of cells. For example, there are stains that specifically stain myelin (Luxol Fast Blue), some that only stain neurons (cresyl violet), and some that stain only the genetic material found in cell nuclei (DAPI).
One of the most influential biological stains was developed in the late 1800s by the Italian anatomist Camillo Golgi. The Golgi stain is a silver-based stain that filled every single part of the neuron, from the dendrites to the axons, turning the cells black. The stain is only taken up by less than 1% of neurons, and because of the low rate of staining, individual neurons could be traced in their entirety and drawn. This stain led to a contentious battle between Golgi and a rival neuroanatomist Santiago Ramon y Cajal over the nature of the connections between neurons.
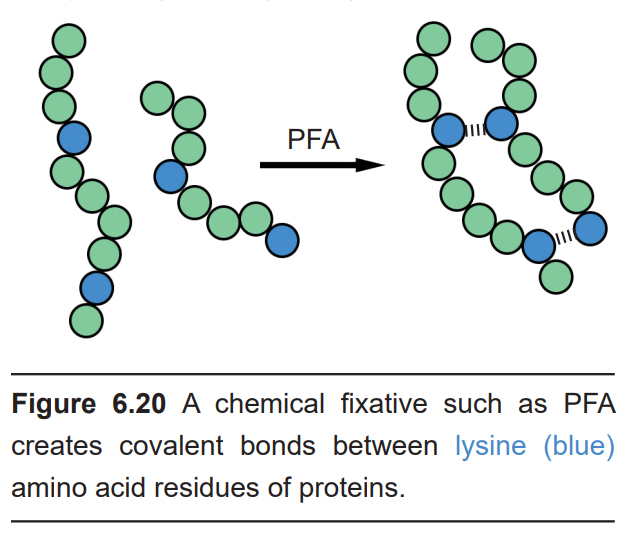
For staining to work, the tissue needs to be subjected to a series of chemical processes. First, the tissue needs to be fixed. Fixation is a chemical process that is accomplished by exposing the tissue to a chemical like paraformaldehyde (PFA). The most effective way to expose every part of the body to fixative is to “hijack” the endogenous circulatory system by flushing fixative through the arteries, a process called perfusion. Chemically, fixatives cause adjacent proteins to become covalently bonded (cross- linked), a process which causes the proteins to become unchangeable – they become “fixed” in time. These fixatives are very harsh chemicals and usually kill microorganisms and inactivate the endogenous enzymes that normally degrade biological tissue. (As a side note, fixatives are particularly nasty carcinogens that can permeate easily through latex gloves.)
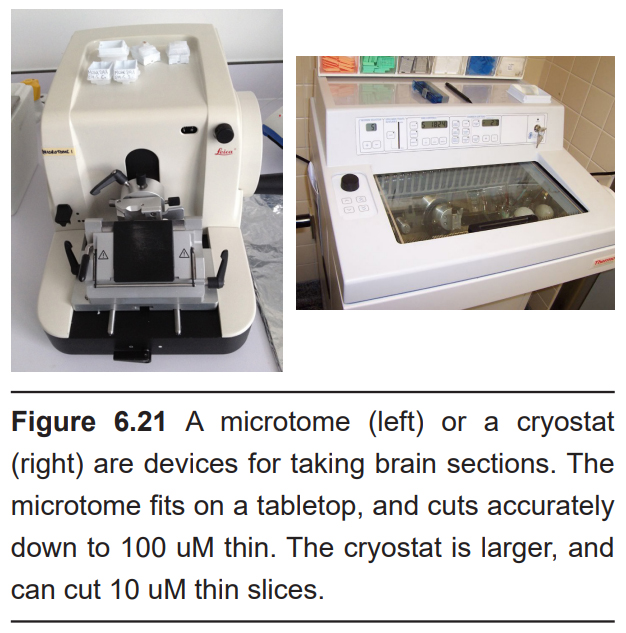
After fixation, devices such as a microtome or cryostat are used to slice the brain into sections as thin as 10 microns. Stains do not always reliably pass all the way through thick sections of tissue such as an intact brain. With thin slices, chemical stains are able to permeate through the depth of the tissue.
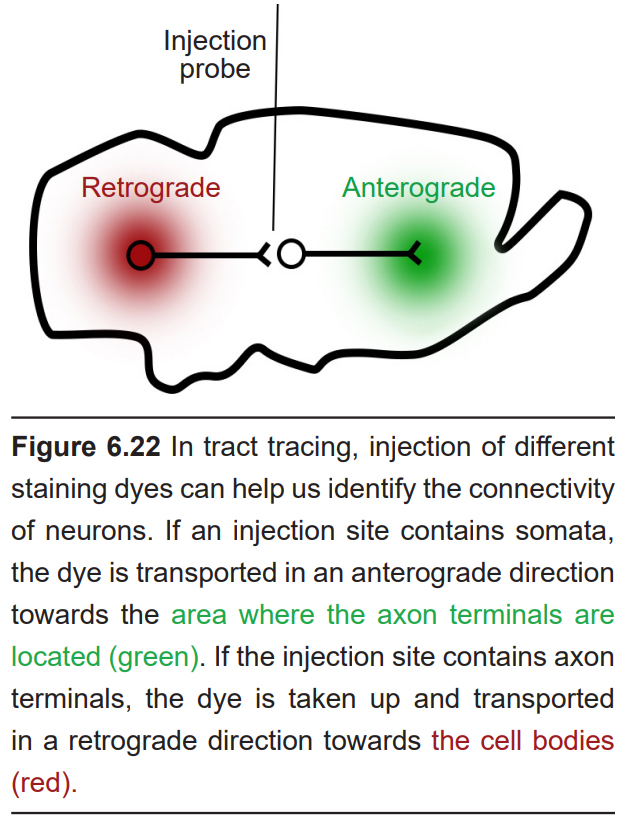
Certain staining methods can also be used to determine projections of neurons. These techniques are collectively called tract tracing methods. An anterograde trace stains cells from the soma to the axon terminal, while a retrograde trace is taken up by the axon terminals and stains the soma. These strategies are helpful for visualizing the nature of connectivity in the nervous system.
Staining cannot be done on living tissue; it can only work on fixed tissue.
6.3.3 Immunohistochemistry staining
Example questions answered:
“Where is the protein actin located in a neuron?” “Is the protein PSD-95 expressed in the same areas as the NMDA receptor protein?”
Immunohistochemistry (IHC) is a modification of the staining methods that gives increased specificity over the other chemical stain methods presented above. IHC is used because it is good for identifying the location of specific proteins at a subcellular level. In other words, you can use IHC to figure out where specific neurons are located inside or on the cell.
As with other staining methods, IHC begins with thinly prepared sections of fixed tissue. Instead of exposing the tissue to various chemical stains, IHC utilizes the molecular properties of antibodies, proteins that function in the immune response. Each antibody has a part of their Y-shaped structure that is able to bind to other targets (which are called antigens) with extremely high specificity. This other target could be a specific protein, such as serotonin receptors or amyloid precursor protein, or an enzyme such as choline acetyltransferase. Antibodies are created with a particular target in mind, so when the antibody is bathed over the brain tissue, the antibody will adhere only to that target.
The IHC protocol often uses two different antibodies. The first antibody, called the primary antibody, is selected because of its ability to bind to the antigen that you are interested in studying. For example, if you wanted to search for the location of a protein called NeuN, the primary antibody would be anti-NeuN. Afterwards, the tissue will be exposed to the secondary antibody.
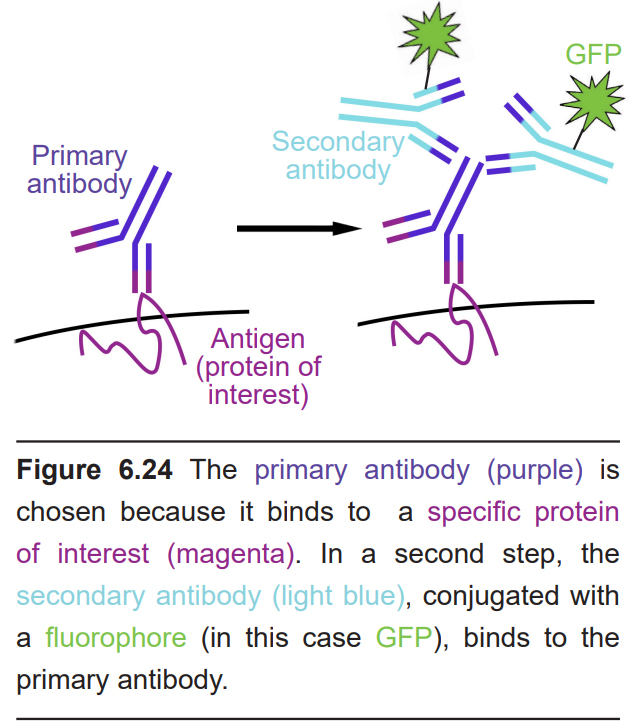
This second antibody is chosen because its target is something that is found on the primary antibody. As an additional feature, the secondary antibody is conjugated with a light sensitive fluorophore, or light-producing molecule. Activation of this fluorophore on the secondary antibody under fluorescence microscopy is then used to identify the location of the primary antibody.
It is also possible to use antibodies and fluorescence microscopy to visualize the elements of cells in a culture dish (in vitro). The theory behind this technique is the same as in IHC, but the name is different. Instead, this approach is called immunocytochemistry (cyto = cell, while histo = tissue). This immunolabeling strategy can also be applied to a whole brain after it has been made transparent through CLARITY (section 6.1.3).
One of the main difficulties with immunochemistry is that the antibodies often exhibit nonspecific binding. Although antibodies are used because of their ability to recognize and differentiate between similar-looking antigens, often they bind to the wrong targets. When they do, it creates a false positive, causing a fluorescent signal to appear when there is actually no protein present. Nonspecific binding also causes an increase in background noise, making it more difficult to identify a genuine fluorescent signal, indicating presence of the protein of interest. Various steps can be taken during the immunochemistry procedure that minimize the likelihood of nonspecific binding, such as thorough rinsing and exposure to blocking agents.
For an immunochemistry protocol to work, an antibody must exist for the specific target of interest. However, an antibody has not been developed against every single protein or enzyme. Another issue is that some proteins are structurally very similar to other proteins, and an antibody has not yet been developed that can differentiate the two. For example, the structure of the dopamine receptor type 2 is so similar to type 3, that there are no antibodies that can target one without the other.
6.4 Changing nervous system activity
Visualizing the gross anatomy or cellular structures of the nervous system allows us to ask very meaningful scientific questions. However, sometimes you are interested in controlling some part of the nervous system, either by increasing or decreasing activity. Using the following neural manipulation strategies, it becomes possible to establish causation between some aspect of the nervous system (neuron, structure, neurotransmitter, gene, etc.) and function.
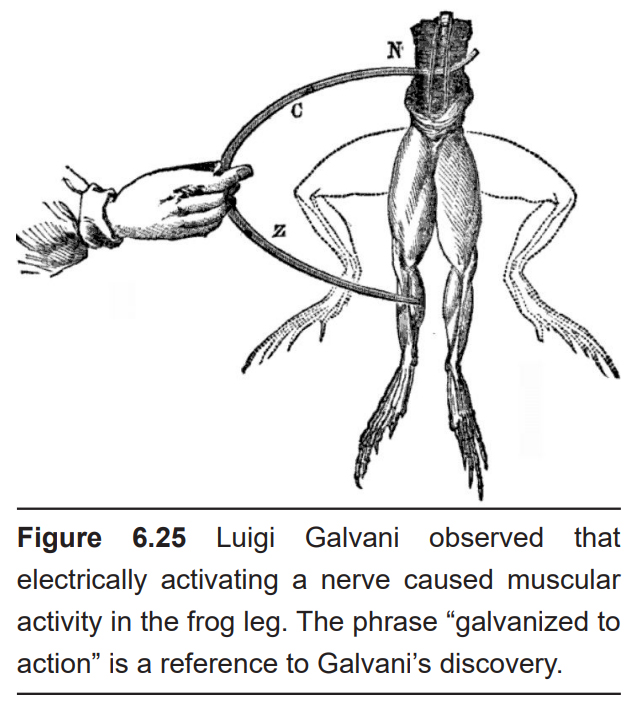
Early neural manipulation strategies trace back to the late 1700s with the accidental discoveries of the Italian biologist Luigi Galvani. With a dissected frog leg on his lab bench, Galvani discovered that he could cause the muscles to twitch by electrically activating the nerves that innervated the muscles of the leg. He called this observation “bioelectricity,” which is the idea that endogenous electrical activity is important for muscle control, and by extension, the activity of the whole organism.
Neural manipulation became possible in humans following the work of the brain surgeon Wilder Penfield. In the 1950s, Penfield worked to develop a treatment for epilepsy. In this procedure, the patient is given a local anesthetic while the skin above their skull was resected, allowing access to the brain. The surface of the brain has no pain receptors, so brain surgery can be done while the patient is completely awake. With the patient’s brain exposed, Penfield used a brief electrical pulse delivered by an electrode placed at the surface of the brain. The stimulation activates descending motor systems, which caused muscle contractions in very specific muscle groups of the patient. For example, stimulating the dorsal most part of the motor cortex caused a twitch of the upper leg and hip muscles. By moving incrementally across the motor cortex, he was able to draw a graphical representation of which areas of the motor cortex controls which muscles.
The techniques presented here are all methods by which we can induce some type of change in nervous system activity.
6.4.1 Electrophysiology (Ephys)
Example questions answered:
“How frequently do neurons in the striatum fire action potentials?”
“Does increasing cortical activity change the excitability of spinal cord neurons?”
Electrophysiology relies on using specialized equipment, informally called a rig, to measure and manipulate the electrical properties of the nervous system. Originally, electrophysiology was used to detect the difference in electrical potential between the inside of the neuron and the outside. The earliest electrophysiology techniques examined the giant axon of the Loligo squid. The axon itself is so large with a diameter around 1 mm, the inside of these neurons are very easy to access by running a conductive probe into the cell. Two researchers, Alan Hodgkin and Andrew Huxley, earned a Nobel prize in 1963 for the work that came out of these electrophysiological techniques.
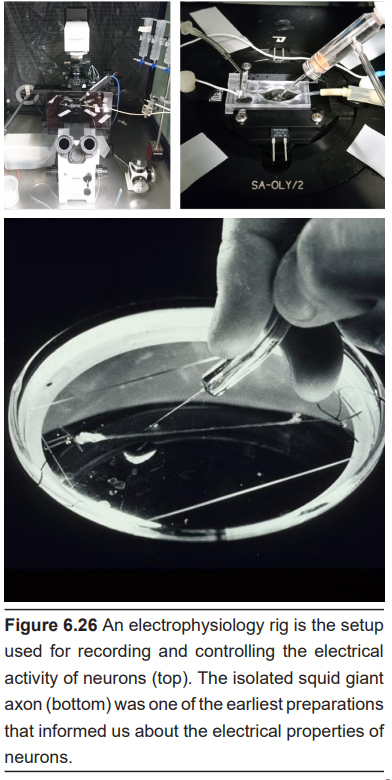
Electrophysiology on a cellular level usually requires the concurrent use of a microscope to get resolution down at the level of the neuron, which may be on the order of tens of microns in diameter. At this level of magnification, tiny glass micropipettes filled with an electrolyte solution can then be inserted into neurons, and electrical currents can then be detected and recorded. These currents can be the currents associated with the movement of charged ions, such as sodium during an action potential.
Electrophysiology can also be used to manipulate the activity of neurons. Instead of simply detecting currents, electrophysiology gives you direct physical control over the electrical properties of the neuron, which gives us the ability to manipulate different components of the nervous system.
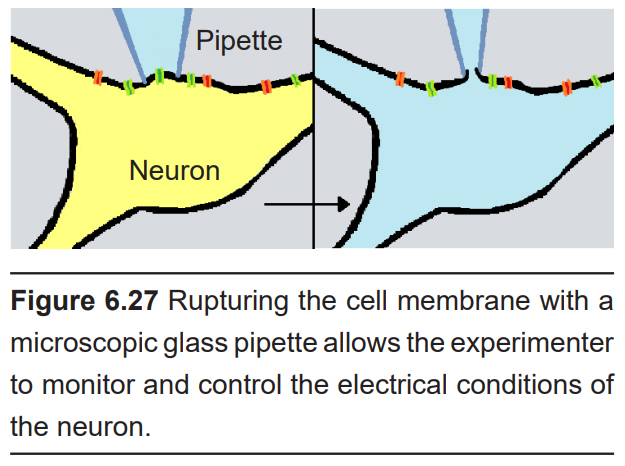
Electrophysiology can be a very versatile technique. Because there are many strategies that count as electrophysiology, you can ask questions at many levels, from behavior and neuronal circuitry to individual neurons and ion channels. With electrophysiology, you can record the patterns of neuronal activity as a rat performs on a behavioral task and stimulate neurons to see if you can modify the behavior. On the other extreme, you can examine the activity of individual ion channels, to determine the conditions when they open and close.
One weakness lies in the highly experimental nature of electrophysiology. There are many different preparations that can be used in studying the electrical activity of the nervous system: intact anesthetized, thin slice of living brain, neuronal culture, or even frog oocytes that express neuronal receptors. But, each step you move away from the awake and behaving animal, the more difficult it becomes to draw inferences based on our results. What we gain in control over the experimental preparation, we lose in ability to generalize about data.
6.4.2 Transcranial magnetic stimulation (TMS)
Example questions answered:
“Which part of the motor cortex sends signals to the contralateral foot?”
“Can activation of parts of the prefrontal cortex decrease the symptoms of depression?”
In physics, there is a close relationship between electricity and magnetism. The movement of magnets can generate electric currents, and conversely, electricity can generate magnetic fields, a process called induction. A brain stimulation technique called transcranial magnetic stimulation (TMS) relies on induction to generate electrical currents through the skull. The TMS machine itself is a handheld coil of wires in a loop. Passing electrical current through that loop produces a magnetic field. The magnetic field generated then induces an electrical current at some distance away from the coil. By placing the coil at the surface of the scalp, we can electrically activate small areas of brain tissue. For instance, using TMS above the motor cortex causes muscle contractions, and using TMS above the occipital lobe causes the perception of flashes of light.
TMS is believed to have moderate benefits for several different psychiatric conditions, potentially making it a meaningful therapeutic intervention. Activation of motor cortex may help alleviate chronic pain conditions, Parkinsonian symptoms, and improve contralateral motor function following stroke injury. Activation of prefrontal cortex can decrease anxiety, antidepressive symptoms, cigarette craving or consumption, and schizophrenia. Stimulation of other brain regions can be antiepileptic, and may also minimize auditory hallucinations or tinnitus (ringing in the ears.)
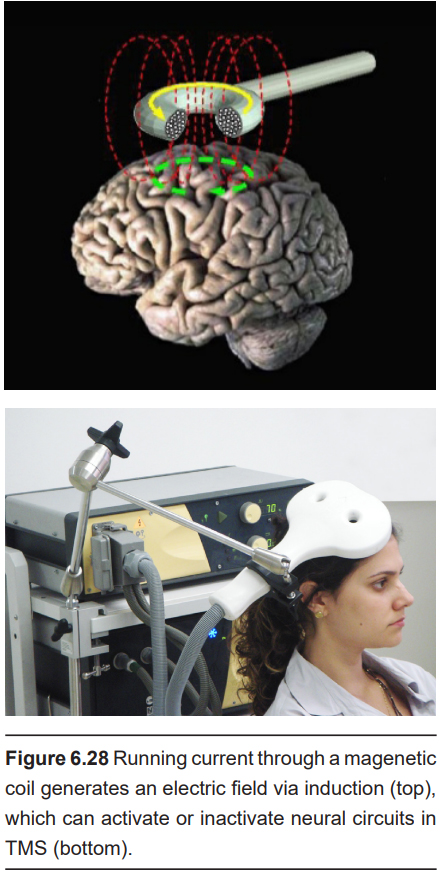
The technique itself is completely noninvasive. TMS can be delivered simply with placement of the electric coil on the surface of the head.
TMS may have some unexpected consequences, such as temporary headaches, localized pain, changes in hearing, and bizarre changes in somatosensation. The most severe side effect so far recorded is seizures, which can be very dangerous. Use of TMS is still highly experimental.
As with fMRI, the magnetic fields generated by a TMS device can cause dangerous interactions with any magnetosensitive implants, such as deep brain stimulation devices, cochlear implants, or aneurysm clips.
Introduction to Computational Neuroscience
Computational neuroscience is a branch of neuroscience that focuses on explaining neural mechanisms through computational methods. This multi-disciplinary field draws from other fields such as electrical engineering, physics, and computer science to better understand how the nervous system processes information. Computational neuroscience strives to answer a wide range of questions in neuroscience research, including computation in single neurons, computation in a neural network, neuron development, sensory processing, motor control, memory and plasticity, as well as broader questions about vision, consciousness, and cognition. Sometimes, using computational approaches can be described as being an in silico preparation.
Computational neuroscience provides tools and methods for developing models, which are used for answering different categories of questions.
- Descriptive models characterize what nervous systems do. These models qualitatively describe data through neural encoding as well as extract information through neural decoding, the reconstruction of information that has already been encoded and represented in the brain by a network of neurons. Reconstruction is the researcher’s ability to predict sensory stimuli purely from the action potentials from neurons. The objective of neural decoding is to characterize the electrical activity of neurons in relation to responses in the brain. Neural encoding characterizes the relationship between the stimulus and the neuronal responses and the relationship with electrical activity.
- Mechanistic models are used to determine how the nervous system relates with known anatomy and physiology. These models simulate the behavior of a single neuron or network of neurons.
- Interpretive models lead to an understanding of why the nervous system operates in a particular way. These models are used to examine neural cognition and the underlying nervous system functionality.
There is debate with how computational models can be used to best reflect the nervous system. A model that focuses largely on defining biological processes into mathematical terms may stray from its intended purpose and not accurately reflect neural behavior. In other cases, a mathematical model may oversimplify neural behavior and may lead to false conclusions. Thus, there is a focus on biologically plausible neural networks to best model the nervous system.
6.4.3 Genetic modification
Example questions answered:
“Do rats behave differently if they do not synthesize the hormone leptin?”
“How do mutated voltage-gated sodium channels change their neuronal action potential firing?”
Since the rise of molecular genetics techniques in the 1970s, researchers have discovered some of the underlying genetic commonalities between healthy people and those with neurological or psychiatric conditions. The genomes of many animals have been completely mapped out including the nematode C. elegans (1998), drosophila (2000), mouse (2002), human (2003), rat (2004), and the macaque monkey (2007).
Knowledge of their genomes, and the capability to manipulate their genomes, has allowed researchers to create animal models of a variety of human conditions. For example, we can use gene excision to remove a small section of the genetic code, such as the part that codes for a certain protein. This strategy is called knock- out, and it is helpful for identifying the function of a genetic sequence. Alternatively, we can use gene insertion to put exogenous genes into an animal. For example, we can generate mice that express the human version of a gene. We call these humanized mice. This type of modification is called a knock-in. There is an entire spectrum of genetic modifications that can be performed, including knock-downs (moderate decrease in function), upregulations (some increase in function), or conditional knock-outs (an organism that is normal until exposed to certain chemicals).
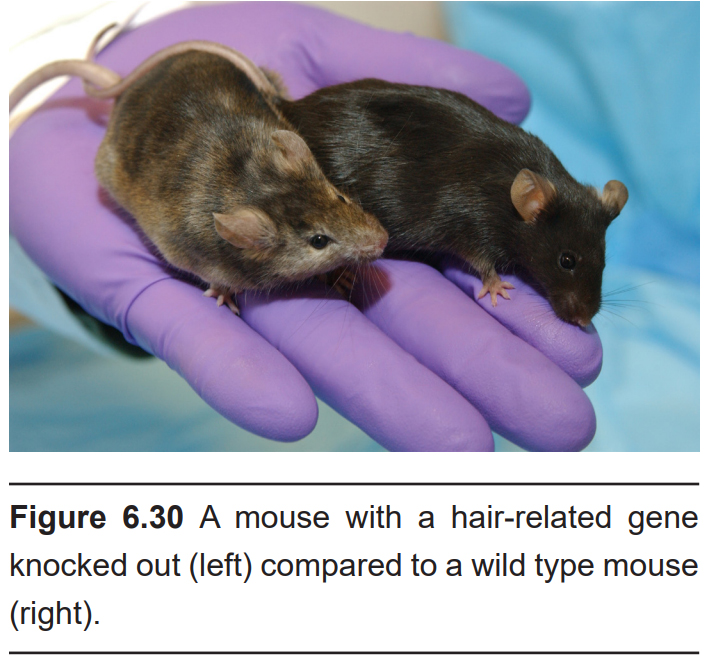
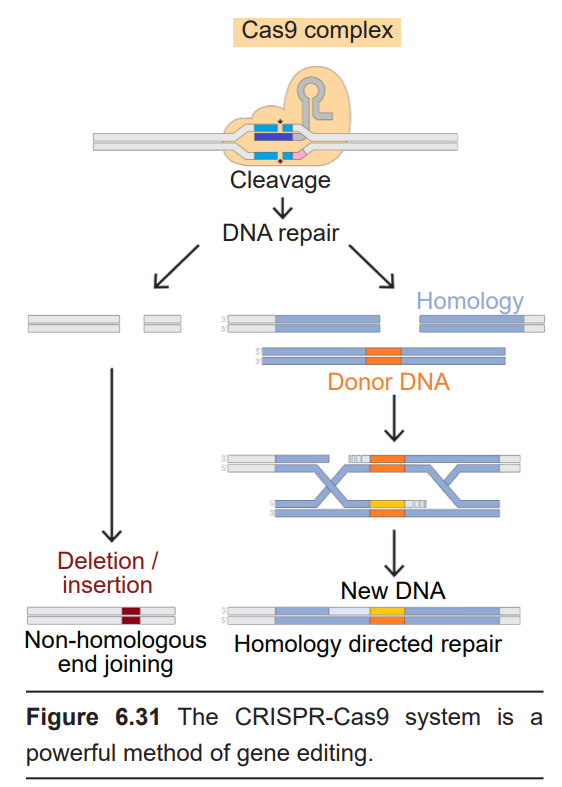
A recent advance in genetic modification technology published in 2012 called the CRISPR- Cas9 system allows for targeted editing of the genome. The main advantage provided by CRISPR-Cas9 is the simplicity, efficiency, and precision of the method. While still highly experimental, there are potential therapeutic applications for CRISPR-Cas9 in treating a variety of genetic diseases.
With the combined help of dedicated laboratories across the world, a wide variety of genetically-modified mice are commercially available for research purposes. Many scientific advances have come from these research models. It’s also possible to develop an animal with multiple genetics crosses, which allow for several complex questions to be answered.
A shortcoming of the information gained from these genetic modification techniques is the difficulty in generalizing the findings beyond the specific genetic strain. For example, consider a knock-in approach to model a brain disorder in mice. If a therapy is developed that helps these animals, it is certainly a starting place that may lead to a human therapy. But, there is no certainty that the therapy will also be effective in humans.
Genetic modifications may also produce unexpected side effects as a result of changes to the genetic code. It is difficult to predict how a specific gene manipulation will interact with other aspects of the animal’s physiology.
6.4.4 Optogenetics / Chemogenetics
Example questions answered:
“How does activating cortical glutamatergic drive affect firing rate of neurons in the striatum?” “Does inactivation of GABA-ergic circuits in the amygdala change anxiety behaviors?”
One of the problems with early neural stimulation strategies was a lack of specificity. For example, many brain regions are home to a hodgepodge of cell populations that produce and release different neurotransmitters. In activating these areas, it was impossible to know if the effect of stimulation was due to activity of a specific population of neurons. Additionally, we were unsure if the results of electrical stimulation were due to the activity of that specific brain area where the stimulator was placed, or if it was due to the activation of axonal fibers that simply pass through that area. This was described as the fibers of passage problem.
The following two strategies, optogenetics and chemogenetics, were recent advances in neuroscience techniques that increase the specificity of brain stimulation. As of now, both of these techniques are exclusively used in nonhumans for research purposes.
Optogenetics
A revolutionary strategy developed in 2006 by researchers from Stanford University, optogenetics use light sensitive-ion channel proteins called channelrhodopsin, or ChR2. The light-sensitive component of ChR2 is a protein that is derived from the rhodopsin molecule, the same proteins that allow photoreceptors in the visual system to detect light. The ion channel component is a cation-permeable transmembrane pore that can allow Na+ ions to move across the cell membrane. When a photon of blue light hits the ChR2 molecule, the protein physically changes shape, causing the cation channel to open, enabling inward flow of positively charged sodium ions. Therefore, ChR2 gives us the ability to excite neurons with flashes of light.
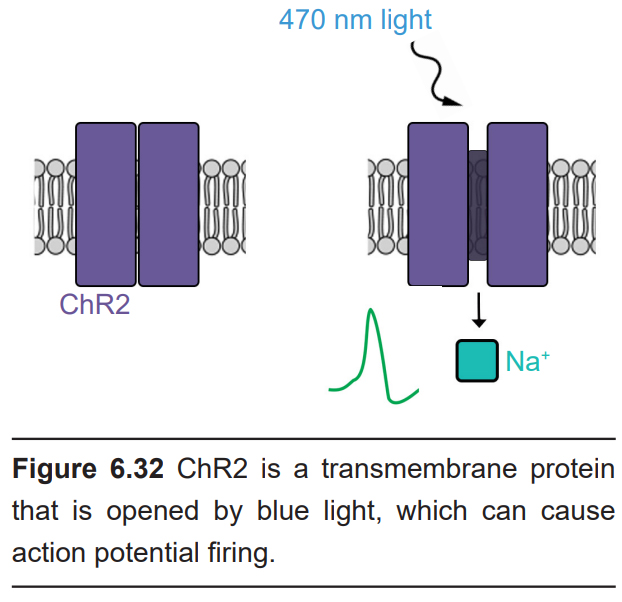
The ability for ChR2 to change structure in response to light is very fast, and the subsequent movement of changed ions down their electrochemical gradient is even faster. A brief 0.5 millisecond flash of blue light is enough to trigger sufficient inward Na+ current to induce a single action potential. Once the blue light is turned off, the channel closes in less than a millisecond. The speed with which ChR2-expressing neurons can respond gives optogenetic strategies a temporal resolution on the order of milliseconds.
ChR2 is possibly the most popular optogenetic molecule, but not the only one. Since its adoption into neuroscience, there have been several other proteins with similar light-sensitive characteristics. Halorhodopsin is a light-sensitive pump that uses energy from yellow-green wavelengths to actively push chloride into the cell, which inhibits neurons. Archaerhodopsin is a light-sensitive proton pump that extrudes H+ ions, which inhibits neurons.
ChR2 also has very high specificity, since optogenetics is used in conjunction with knock-in genetic modification technology or viral gene delivery to insert ChR2 into unique cell populations. This strategy allows researchers to isolate the effect of activation of a specific cell population within an area of the brain that has many different cell types.
Chemogenetics
Chemogenetics is an alternative strategy to selectively activate specific cell populations. Instead of using light to activate special receptors, these techniques use various chemical agonists. Chemogenetic receptors are usually G-protein coupled receptors that are activated only by exogenous drugs, but not by any endogenous neurotransmitters. The most well-characterized chemogenetic receptor is called DREADD, which stands for designer receptor exclusively activated by designer drugs. When the receptor is exposed to the designer drug, it can trigger the activation of the intracellular signaling pathway that can either excite or inhibit the neuron, depending on the nature of the DREADD. So, when a DREADD protein is inserted into a neuron, there are no changes in neuronal activity at rest. But once the receptor is exposed to the endogenous drug, the DREADD is activated, which changes the activity of the neuron.
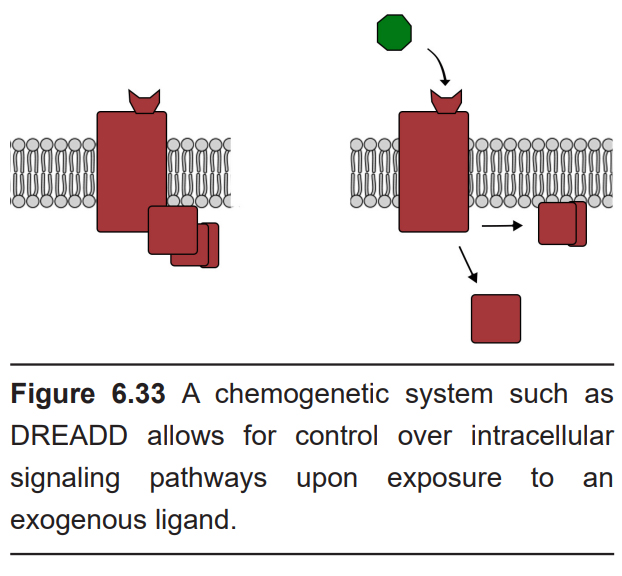
Because chemogenetics requires the drug to enter the system and subsequent GPCR activation before a change in behavior can be observed, DREADD offers significantly less temporal resolution than optogenetics (in an ex vivo preparation – seconds to tens of seconds; in a behaving animal – minutes).
Optogenetics / chemogenetics can be used simultaneously with other techniques. Combining optogenetics with electrophysiology techniques has taught neuroscience about the nature of the connectivity between brain areas. Behavioral testing with simultaneous optogenetic or chemogenetic stimulation has given us insight into the roles that are played by specific neural circuits or neurotransmitters. These techniques are quickly advancing our understanding of the nervous system, and will continue to do so in the future.
Image Credits
Cover: https://upload.wikimedia.org/wikipedia/commons/f/fb/Brain_MRI.jpg
6.1: Image by Pete Linforth from Pixabay https://pixabay.com/photos/countryside-tree-landscape-sunlight-2175353/
6.2: https://pixabay.com/photos/ara-parrot-yellow-macaw-bird-3601194/
6.4: https://upload.wikimedia.org/wikipedia/commons/c/cc/Adult_Caenorhabditis_elegans.jpg https://commons.wikimedia.org/wiki/File:Standing_female_Drosophila_melanogaster.jpg; https://commons.wikimedia.org/wiki/File:Mouse_white_background.jpg https://pixabay.com/photos/rat-garden-out-curious-sweet-1379317/; https://pixabay.com/photos/animal-monkey-3892155/; https://pixabay.com/photos/personal-network-social-media-3139194/
Experimental preparations: https://commons.wikimedia.org/wiki/File:54986main_mouse_med.jpg https://upload.wikimedia.org/wikipedia/commons/c/c7/Galvani-frogs-legs-electricity.jpg; https://upload.wikimedia.org/wikipedia/commons/0/05/Cell_culture.jpg
6.5: https://upload.wikimedia.org/wikipedia/commons/f/fe/M%C3%BCller-Lyer_illusion.svg
6.6: https://wellcomecollection.org/works/vyfjw978?wellcomeImagesUrl=/indexplus/image/L0028667.html
https://commons.wikimedia.org/wiki/File:1895-Dictionary-Phrenolog.png
6.7: https://commons.wikimedia.org/wiki/File:Medical_X-Ray_imaging_WYJ07_nevit.jpg
6.8: https://pixabay.com/photos/bethesda-naval-medical-center-80380/
6.9: https://commons.wikimedia.org/wiki/File:Ct-scan_of_the_brain_with_an_subdural_hematoma.jpg modified by Austin Lim
6.10: https://upload.wikimedia.org/wikipedia/commons/0/09/The_diffusion_tensor_tractographies_of_neural_tracts_for_ language_fnhum-07-00749-g001.png
6.11 : https://pubs.niaaa.nih.gov/publications/arh27-2/146-152.htm modified by Austin Lim
6.12 : Optimization of CLARITY for Clearing Whole-Brain and Other Intact Organs Jonathan R. Epp, Yosuke Niibori, Hwa- Lin (Liz) Hsiang, Valentina Mercaldo, Karl Deisseroth, Sheena A. Josselyn, Paul W. Frankland eNeuro 8 May 2015, 2 (3) ENEURO.0022-15.2015; DOI: 10.1523/ENEURO.0022-15.2015
6.13 : https://upload.wikimedia.org/wikipedia/commons/b/bf/EEG_cap.jpg; https://upload.wikimedia.org/wikipedia/commons/f/f1/Gen-20s_SWK.png modified by Austin Lim
6.14: https://commons.wikimedia.org/wiki/File:Fluorodeoxyglucose_18-F_skeletal.svg
6.15: https://upload.wikimedia.org/wikipedia/commons/7/7b/PET_scan.jpg https://upload.wikimedia.org/wikipedia/commons/2/2a/Pet_scan_of_brain_tumor.jpg modified by Austin Lim
6.17: https://upload.wikimedia.org/wikipedia/commons/2/2a/FMRI_BOLD_activation_in_an_emotional_Stroop_task.jpg modified by Austin Lim
6.18: https://upload.wikimedia.org/wikipedia/commons/e/e1/FluorescenceFilters_2008-09-28_cs.svg modified by Austin Lim
https://upload.wikimedia.org/wikipedia/commons/9/9e/DGCs_in_GFP.jpg
6.19: https://upload.wikimedia.org/wikipedia/commons/5/5e/Cerebellum_Cross_Section_Medulla_%2842040992452%29. jpg; https://upload.wikimedia.org/wikipedia/commons/6/6d/GolgiStainedPyramidalCell.jpg
6.21: https://upload.wikimedia.org/wikipedia/commons/9/9c/Cryostat_microtome.jpg
https://upload.wikimedia.org/wikipedia/commons/4/46/Leica_microtome_2.JPG
6.23: https://upload.wikimedia.org/wikipedia/commons/d/dc/PLoSBiol4.e126.Fig6fNeuron.jpg
https://upload.wikimedia.org/wikipedia/commons/0/09/FluorescentCells.jpg
6.25: https://upload.wikimedia.org/wikipedia/commons/c/c7/Galvani-frogs-legs-electricity.jpg
6.26: https://upload.wikimedia.org/wikipedia/commons/1/13/Patch_rig.png; https://upload.wikimedia.org/wikipedia/commons/1/19/Giant_Axon_of_Squid_%2814356033761%29.jpg
6.27: https://upload.wikimedia.org/wikipedia/commons/9/9c/Whole_Cell_Patch_Clamp_Steps.png modified by Austin Lim 6.28: https://upload.wikimedia.org/wikipedia/commons/6/67/Transcranial_magnetic_stimulation.jpg; https://upload.wikimedia.org/wikipedia/commons/3/38/Neuro-ms.png
6.29: https://upload.wikimedia.org/wikipedia/commons/5/5f/Brain-Controlled_Prosthetic_Arm_2.jpg
6.30: https://upload.wikimedia.org/wikipedia/commons/e/e1/Knockout_Mice5006-300.jpg
6.31: https://upload.wikimedia.org/wikipedia/commons/0/04/DNA_Repair-colourfriendly.png Modified by Austin Lim
The Open Neuroscience Initiative is funded by a grant from the Vincentian Endowment Fund of DePaul University.
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.

